All published articles of this journal are available on ScienceDirect.
Biological Evaluation and Reverse Pharmacophore Mapping of Innovative Bis-Triazoles as Promising Anticancer Agents
Abstract
Here, we describe further cytotoxic studies and reverse pharmacophore mapping (pharmacophore profiling) for bis-triazoles MS44-53, which were designed and synthesized previously to stabilize the G-quadruplex nucleic acids capable of being formed at the telomeric region and promoter sequences of genes involved in cellular proliferation and oncogenes. Pharmacophore-based activity profiling screen demonstrated some biological targets that MS44-53 may modulate their biological response, and thus can be considered as potential drugs to treat different kinds of diseases, such as carcinoma, diabetes type II, bacterial infection and cardiovascular diseases. Potent cell growth inhibitory properties were shown by ligands MS47 and MS49 against human melanoma MDA-MB-435, colon cancer HCT-116 and COLO 205, and pancreatic cancer MIA PaCa-2 cell lines, as evidenced by MTT assay. Both ligands were more potent against cancer cells than in skin normal CCD-1064Sk fibroblasts.
Aim:
The aim of this study is to identify the molecular target and mechanism of action of our promising anticancer bis-triazoles MS44-53, focusing specifically on the G-quadruplex stabilizers MS47 and MS49.
Background:
In molecular biology, G-quadruplexes (also known as G4-DNA), one of the higher-order structures of polynucleotides, are four stranded structures formed by nucleic acid sequences which are rich in guanine. They are formed mainly at the single-stranded G-overhang of telomeric DNA and within promoter sequences of genes involved in cellular proliferation and oncogenes such as c-myc, c-kit, and Hsp90. Stabilization of DNA G-quadruplexes is one of the anticancer strategies that has the potential to treat all cancers regardless of the type. A new series of bis-triazoles MS44-53 were developed to stabilize G-quadruplex structures selectively, as G4 ligands and experimental antitumour agents. FRET assay showed that MS47 and MS49 were only the best binders towards the Hsp90 promoter G-guadruplexes. While all bis-triazoles MS44-53 exhibited potent cell growth inhibitory activity against human carcinoma cell lines, suggesting that the ligands perturb molecular targets and mechanisms of action, other than stabilizing G-quadruplexes, contributing to antitumor activity. Therefore, the molecular targets and mechanisms of action of bis-triazoles MS44-53 in different types of human cancer cell lines should be determined by performing further computational studies to MS44-53 and in vitro evaluations for the G-quadruplex stabilizers MS47 and MS49.
Objectives:
1- Determining the exact IC50 for bis-triazoles MS47 & MS49 against four different types of human cancer cell lines; melanoma MDA-MB-435, pancreatic cancer MIA PaCa-2, and colon cancer HCT-116 and COLO 205 cell lines.
2- Predicting the biological targets that bis-triazoles MS44-53 may interact with to trigger or block their biological response.
Methods:
1- MTT assay was used for in vitro evaluation of the antiproliferative activities of MS47 and MS49, and determination of IC50 values.
2- Reverse pharmacophore mapping (pharmacophore profiling) was used for predicting the biological targets of bis-triazoles MS44-53, and determining the % binding probabilities.
Results:
MS49 exhibited more potent proliferation inhibitory activity than MS47 and higher IC50 value against skin normal fibroblasts. Pharmacophore profiling demonstrated FGFR1, PDGFR2, FLT3, mTOR, PPAR-gamma, MUR-F and CETP as biological targets for bis-triazoles MS44-53.
Conclusion:
Bis-triazoles MS47 and MS49 are promising selective innovative compounds with wide spectrum cytotoxic activities against distinct cancer types. Bis-triazoles MS44-53 can be considered as potential drugs to treat different types of carcinoma, in addition to diabetes type II, bacterial infection and cardiovascular diseases.
Other:
Further in vitro evaluations will be performed for bis-triazoles MS44-53 in order to identify their molecular targets and mechanisms of action in different types of human cancer cell lines.
1. INTRODUCTION
Cancer is a group of diseases characterized by the uncontrolled growth and spread of abnormal cells. If the spread is not controlled, it can result in death [1, 2]. In EU-27, the estimated new cancer cases were approximately 1.4 million in males and 1.2 million in females, with over 710,000 estimated cancer deaths in males and 560,000 in females in 2021 [3]. The earliest method of treating cancer was surgery or physical removal of the affected cells and tissue, and this remains the primary initial treatment for several cancers today, including those of the breast, colon and prostate [4]. The success of surgery is often enhanced by the use of additional therapies that are aimed at killing cancer cells which may remain in the body, such as chemotherapy and radiation therapy [5]. Increasingly, specific or “targeted” cancer therapies are being used to increase survival rates and limit side effects that can severely impact the quality of life. These include biological therapies such as vaccines and monoclonal antibodies, which can be used alone or in conjunction with the traditional treatments of surgery, chemotherapy or radiation therapy [6, 7]. Investigation into the design and synthesis of small molecules to interact with proteins, enzymes or components of signalling pathways that are unique or overexpressed in cancer has been ongoing for many years [8]. However, adverse side effects and drug resistance disrupt most of the cancer therapies and motivate research to develop novel anticancer strategies that target and overcome this prevalent and often fatal disease [9, 10].
Stabilization of DNA G-quadruplex (also known as G4-DNA) is one of the anticancer strategies that has the potential to treat all cancers regardless of type [11]. It has been suggested that ligands that bind selectively and stabilise G4-DNA at the telomeric region or the promoter sequences of genes involved in cellular proliferation and oncogenes such as c-myc, c-kit, and Hsp90 in cancer cells, could result in senescence (growth arrest) or delayed cell death, and abrogation of tumorigenicity [12-15].
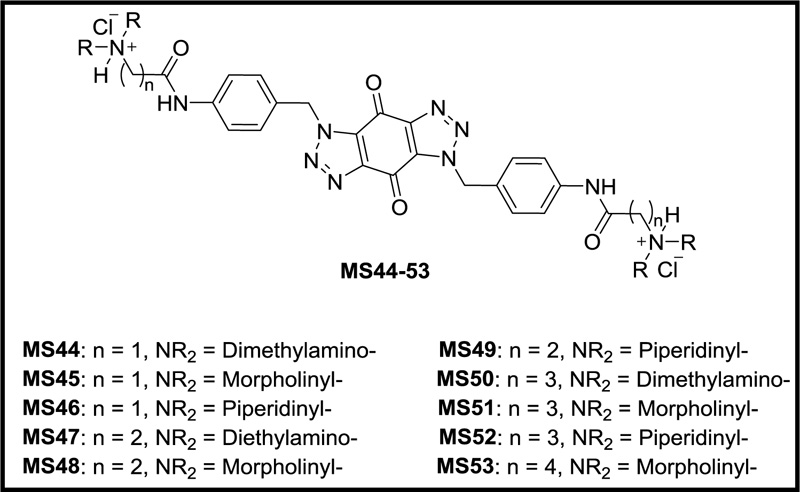
Previously, we designed and synthesized a new series of ten novel bis-triazoles, MS44-53 (Fig. 1), to stabilize G-quadruplex structures selectively as G4 ligands and experimental antitumor agents [16]. FRET assay [17] indicated that ligands MS47 and MS49 were the best binders towards the telomeric and Hsp90a promoter G-quadruplexes, with selectivity towards the latter. Compared with the known G-quadruplex binder RHPS4, both ligands can be considered G4 moderate binders [16]. In MTT antiproliferative assay [18], all the ligands, especially binders MS47 and MS49, showed potent cell growth inhibitory activity against human carcinoma (colon and pancreatic) cell lines and good selectivity for them over the human normal lung fibroblasts cell line, making them promising antitumor agents for cancer therapy [16].
Ligand MS47 was sent to NCI (National Cancer Institute) [19], Washington, USA, to be evaluated biologically against 60 human epithelial cancer cell lines using MTT assay. The mean graph resulting from NCI 60 Cell One-Dose Screen indicated that MS47 (named as NSC778438) satisfied predetermined threshold inhibition criteria in a minimum number of cell lines. Furthermore, ligand MS47 was advanced to the NCI 60 Cell Five-Dose Screen and reported mean graphs of three levels of effect: 50% Growth Inhibition (GI50), Total Growth Inhibition (TGI), and 50% Lethal Concentration (LC50). The data and mean graphs indicated that MS47 has selective, potent cell growth inhibitory and cytotoxicity against different types of tumour cell lines, making it a promising drug for various carcinoma diseases, such as melanoma, colon, CNS, ovary, breast, and renal cancers.
Herein, we describe the anticancer activity of MS47 and MS49 against different types of human cancer cell lines such as melanoma MDA-MB-435, colon HCT-116 and COLO 205, and pancreatic MIA PaCa-2 cell lines, and their selective cytotoxicity to cancer cells over normal skin fibroblasts, using MTT assay. Furthermore, we report the biological targets that bis-triazoles MS44-53 may initiate or inhibit their biological response, predicted by reverse pharmacophore mapping.
2. MATERIALS AND METHODS
2.2. Cell Culture
Human melanoma MDA-MB-435, pancreatic carcinoma MIA PaCa-2, colorectal carcinoma HCT-116, colorectal adenocarcinoma COLO 205, and skin normal fibroblast CCD-1064Sk cell lines were purchased from American Type Culture Collection (Manassas, VA, USA) (ATCC), USA and stored in liquid nitrogen. Cells were passaged twice weekly upon reaching 70–80% confluency, and low passage numbers (≤ 20) were used in all experiments. Lonza MycoAlert™ mycoplasma detection kits were used (according to the manufacturer's instructions), ensuring cultures were contamination-free. Cells were sub-cultured in RPMI 1640 medium (Sigma-Aldrich (UK), CN: R8758) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco, CA, USA), 100 units/mL penicillin, and 100 μg/mL of streptomycin (Gibco 15140-122). Cells were maintained at 37 °C in a humidified 5% CO2 incubator (Shel Lab., Cornelius, OR, USA).
2.3. MTT Assay: Measuring Cell Viability
The method has been described previously by Saleh et al. [16] and was adapted from Mosmann [18]. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] reagent was used to evaluate the antiproliferative activity of ligands MS47 and MS49 on the human melanoma MDA-MB-435, pancreatic carcinoma MIA PaCa-2, colorectal carcinoma HCT-116, colorectal adenocarcinoma COLO 205, and skin normal fibroblast CCD-1064Sk cell lines. Cells were seeded into 96-well plates (Nunc, Roskilde, Denmark) at the density of 3 × 103 per well (180 µL per well) and allowed to adhere for 24 h at 37 °C/5% CO2. Agent top stock solutions (10 mM in DMSO) were then freshly made. Serial dilutions were prepared in RPMI 1640 medium for addition to carcinoma cells. Control wells received vehicle alone (20 µL per well). Final test agent concentrations in the wells were; 0.01, 0.05, 0.1, 0.5, 1, 5, 10, 50, and 100 µM. The final concentration of DMSO in the wells never exceeded 1%. Vehicle control assays were performed (0.0001–1% DMSO). Experimental plates were incubated for a further 72 h period at 37 °C/5% CO2. Cell viability was recorded at the time of agent addition (T0 ) and after 72 h exposure: following the addition of MTT solution (2 mg mL-1 in PBS: 50 µL per well), experimental plates were incubated for 3 h to allow reduction of MTT by viable cells to insoluble dark purple formazan crystals. The supernatant in each well was then aspirated, and the cellular formazan was solubilized by the addition of DMSO (150 µL per well). Absorbance (OD) was read at a wavelength of 550 nm by using an Anthos Labtec system plate reader. The measured intensity is proportional to metabolic activity, which correlates with viable cell number. Estimated IC50 values (test agent concentrations that inhibit cell growth by 50%) were calculated using Prism (version 7.0) software. Results are expressed as the mean of three independent experiments (n = 8 per trial).
2.4. Reverse Pharmacophore Mapping
The method applied for this study was adapted from Schuster [20]. Bis-triazoles MS44-53 were profiled against an in-house established pharmacophore database. The database consists of a collection of 106 pharmacophore models. The pharmacophore model collection (PMC) reflects different binding modes of 34 drug targets.
The PMC was established in the Molecular Modeling Research Laboratory at the University of Jordan, and the pharmacophores were created mainly by the application of the DiscoveryStudio® (version 4.5) software suite from Accelrys Inc. (www.accelrys.com). Each one of the pharmacophore models has been evaluated individually for its predictive power, using the receiver operating characteristic - area under the curve (ROC-AUC), and all have an AUC ≥ 0.8. The ligands were profiled using the automated Ligand Profiler protocol in the DiscoveryStudio® (version 4.5) software, and mapped to all 106 pharmacophores in the PMC database. The output of profiling includes fit values (0 to 1) of each molecule profiled against each respective pharmacophore. The expressed fit value predicts the activity profiles of ligands. The higher fit value is indicative of the bioactivity of the ligand against a drug target represented by its respective pharmacophore(s) model.
2.5. PMC Compilation
The PMC consists of both ligand based and structure based pharmacophores. Ligand based models were constructed by following a systematic method that includes the following steps: Collecting ligands known to be active against a specified target and grouping them into training sets. Exploring the pharmacophoric space of the training sets and automatically generating pharmacophore models with the application of CATALYST-HYPOGEN module of DiscoveryStudio®. The generated pharmacophores undergo QSAR analysis to nominate the best possible pharmacophore models capable of explaining bioactivity of the training sets. The models’ quality is evaluated using receiver operating characteristic-area under the curve (ROC-AUC). The validation step defines the pharmacophore model’s ability to successfully classify a long list of external compounds (known actives and structurally similar decoys) as actives or inactives. Subsequent addition of steric exclusion spheres is applied using HIPHOP-REFINE module of DiscoveryStudio® to define the steric constraints of the binding pocket and to enhance the quality of the models [21-25].
As for structure-based pharmacophore models, the method starts with the collection of known bioactive molecules against a certain target. These molecules are docked in the protein structure obtained from the Protein Data Bank, and then the docked poses are scored. Docking and scoring are performed using the different docking and scoring engines found in DiscoveryStudio® software. After verifying the best docking configuration, Docking-based comparative intermolecular contacts analysis (dbCICA) is applied. Pharmocophore models are then manually built according to the critical points of ligand-protein interaction that were determined by dbCICA. The models’ quality is validated using ROC-AUC and exclusion spheres are added, just like in the ligand-based pharmacophore models [21, 22, 26-28].
3. RESULTS AND DISCUSSION
3.1. Cell Culture Studies
The antiproliferative activities of MS47 and MS49 were evaluated in vitro using the MTT anti-proliferative assay [18, 29] against MDA-MB-435 (human carcinoma), MIA PaCa-2 (pancreatic carcinoma), HCT-116 (colorectal carcinoma), and COLO 205 (colorectal adenocarcinoma) cell lines. In addition, selective cytotoxicity for MS47 and MS49 was investigated utilizing CCD-1064Sk (human skin normal fibroblast) cell line. Estimated concentrations at which cell proliferation is inhibited by 50% (IC50) were calculated from dose-response curves after 72 h exposure of the cells to MS47 and MS49. The IC50 values are presented in Table 1, while dose-response curves are shown in Fig. (2>).
MS47 and MS49 show potent proliferation inhibitory activities against human carcinoma and normal fibroblast cell lines in the concentration range (0.01-100 µM) used. However, MS49 exhibits more potent proliferation inhibition activity than MS47 in human melanoma MDA-MB-435, pancreatic cancer MIA PaCa-2, colon cancer HCT-116 and COLO 205 cell lines. Notably, melanoma MDA-MB-435 cell line is the most sensitive cell line to ligand MS49, scoring the smallest nanomolar IC50 value of 115.2 nM.
| Human Cell Line | IC50 (nM) ± S.D. | |
| MS47 | MS49 | |
| MDA-MB-435 | 353.4 ± 26.7 | 115.2 ± 28.0 |
| MIA PaCa-2 | 836.9 ± 116.3 | 344.9 ± 52.1 |
| HCT-116 | 466.3 ± 22.8 | 195.4 ± 7.4 |
| COLO 205 | 337.6 ± 37.2 | 195.3 ± 13.4 |
| CCD-1064Sk | 795.0 ± 77.6 | 870.7 ± 62.3 |
To investigate the safety profile of MS47 and MS49, their cytotoxic effect against non-cancerous cells was assayed with the use of CCD-1064Sk (human skin normal fibroblast) cell line. Ligand MS47 exhibits a higher IC50 value against non-cancerous cells in comparison to its IC50 values against melanoma MDA-MB-435, colon cancer HCT-116 and COLO 205 cell lines, with cancer selectivity indexes (SI) of 2.3, 1.7 and 2.4, respectively. While ligand MS49 shows a higher IC50 value against skin normal fibroblasts in comparison to all tested human carcinoma cell lines; MDA-MB-435, MIA PaCa-2, HCT-116 and COLO 205, with SI of 7.6, 2.5, 4.5 and 4.5, respectively.
The results demonstrate that both ligands are potent cytotoxic compounds, however, MS49 has higher potency than MS47 against human carcinoma cells and less toxicity (better safety margin) in human normal cells. Consequently, further biological activity evaluations should be performed for ligand MS49.
3.2. Reverse Pharmacophore Mapping (Pharmacophore Profiling)
Pharmacophore is the spatial arrangement of features essential for a molecule to interact with a specific molecular target. Pharmacophore profiling is a method that can be used as an alternative to molecular docking in order to screen a ligand and find if it is capable of interacting with a potential target [30].

| Ligand | % Probability of Binding (%) | ||||||
| FGFR1 | PDGFR2 | FLT3 | mTOR | PPAR-gamma | MUR-F | CETP | |
| MS44 | 50 | 66 | 72 | 61 | 71 | 65 | 54 |
| MS45 | 56 | 66 | 69 | 61 | 72 | 64 | 50 |
| MS46 | 71 | 64 | 71 | 59 | 71 | 64 | 66 |
| MS47 | 46 | 65 | 66 | 62 | 72 | 66 | 77 |
| MS48 | 52 | 65 | 70 | 61 | 71 | 65 | 65 |
| MS49 | 57 | 64 | 77 | 60 | 71 | 65 | 72 |
| MS50 | 54 | 65 | 63 | 60 | 71 | 65 | 57 |
| MS51 | 58 | 65 | 65 | 60 | 71 | 63 | 61 |
| MS52 | 52 | 65 | 56 | 59 | 72 | 66 | 51 |
| MS53 | 49 | 64 | 71 | 60 | 71 | 66 | 47 |
Bis-triazoles MS44-53 were screened by pharmacophore-based activity profiling screen [20, 31], using Accelrys DiscoveryStudio® 4.5 software, in order to predict their affinity to biological targets. The profiling screen presents the results as percentages of affinity of ligands to a target protein and reflects the probability of their inhibitory or induction capabilities towards these targets. The ligand structure with % binding probability ≥ 50% is believed to fit the binding pocket of the biological target and thus be very close to the proper pharmacophore. The % probability of binding of bis-triazoles MS44-53 to different biological targets; FGFR1, PDGFR2, FLT3, mTOR, PPAR-gamma, MUR-F and CETP, are shown in Table 2.
The results demonstrate noteworthy percentages of binding probability for MS44-53 towards the kinases involved in many cancer diseases, FGFR1, PDGFR2, FLT3, and mTOR, with ranges of 46-71%, 64-66%, 56-77% and 59-61%, respectively, making them promising novel anticancer drugs for the treatment of various carcinoma diseases. MS44-53 showed an interesting % binding probability (71-72%) to the nuclear receptor PPAR-gamma that increases insulin sensitization and enhances glucose metabolism, making them potential agonist hypoglycaemic drugs for treating diabetes type II disease. An antibacterial activity is predicted from the interesting % inhibition probability values (63%-66%) shown by our ligands to MUR-F, the key enzyme in bacterial cell wall biosynthesis in both gram-positive and gram-negative bacteria, making them potential antibacterial agents. Furthermore, the data show a significant % inhibition probability (47% to 77%) for MS44-53 to the plasma lipid transfer protein CETP that facilitates the transport of cholesteryl esters and triglycerides between lipoproteins, making the ligands potential drugs for treating cardiovascular diseases caused by LDL cholesterol.
CONCLUSION
Based on the observed cytotoxic activities of bis-triazoles MS47 and MS49, we present two promising selective innovative compounds with wide spectrum cytotoxic activities against different cancer types. In MTT assays, MS49 showed the most potent cell proliferation inhibitory activity against human melanoma MDA-MB-435 cells, demonstrating selectivity (enhanced potency) over skin normal fibroblasts. Furthermore, human pancreatic cancer MIA PaCa-2, colon cancer HCT-116, and COLO 205 cell lines were more sensitive to MS49. Cancer-selectivity indexes indicated a better safety margin for MS49 than ligand MS47. Pharmacophore profiling predicted that bis-triazoles MS44-53 can be promising anticancer agents via inhibiting FGFR1, PDGFR2, FLT3 and mTOR kinases, hypoglycaemic drugs via binding to the nuclear receptors PPAR gamma, antibacterial drugs through inhibiting MUR-F enzyme, and potential drugs for treating cardiovascular diseases through binding to CETP protein.
LIST OF ABBREVIATIONS
| CCD-1064Sk | = Human skin normal fibroblast cell line |
| COLO 205 | = Colorectal adenocarcinoma |
| GI50 | = 50% Growth inhibitory concentration |
| HCT-116 | = Human colorectal carcinoma cell line |
| Hsp90 | = Heat shock protein 90 |
| Hsp90a | = Labelled G-quadruplex forming oligomer of the promoter sequence Hsp90a oncogene |
| IC50 | = Concentration that inhibited cell growth by 50% |
| LC50 | = 50% Lethal concentration |
| MDA-MB-435 | = Human melanoma cell line |
| MiaPaCa-2 | = Human pancreatic carcinoma cell line |
| MTT | = 3-(4,5-Dimethylthiazol-2-yl)-25-diphenyltetrazolium bromide |
| NCI | = National Cancer Institute |
| OD | = Optical density |
| SI | = Selectivity index |
| TGI | = Total growth inhibition |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The authors would like to acknowledge Isra University in Amman, Jordan, for the financial support under Deans' Council Decision No. 51/3-2018/2019, date of Grant: November 18th, 2018.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Charlie A. Laughton, Prof. Christopher J. Moody and Dr. Tracey D. Bradshaw from University of Nottingham, Nottingham, UK, for providing us with bis-triazoles MS44-53, and Isra University, Amman, Jordan, for the facilities and financial support.


