All published articles of this journal are available on ScienceDirect.
In Silico and In Vitro Evaluation of Bioactive Compounds of Vallisneria spiralis L. against Candida albicans
Abstract
Background
This study focused on the extraction, isolation, characterization, and molecular docking of phytochemicals from Vallisneria spiralis L. to determine its antifungal activity. Natural products and structural analogs have always played an important role in pharmacotherapy, particularly in the treatment of infections and malignancies.
Methods
Vallisneria spiralis L. leaves were pulverized into a fine powder and extracted using a Soxhlet apparatus, and the crude chloroform extract was separated by flash column chromatography. Thin-layer chromatography was used to assess the obtained fractions. Infrared spectra were obtained using a Perkin Elmer spectrometer. NMR spectra were collected using a Bruker spectrometer. Qualitative phytochemical investigations were performed to identify compounds in the extracts. The susceptibility of the test compound to Candida albicans was assessed using disk diffusion. A docking study was conducted to dock the three-dimensional structures of the extracted phytocompounds of Vallisneria spiralis L. into the receptor site of the target proteins using Autodock software. MD simulations were used to study the dynamic behavior, conformational changes, and stability of the proteins using the top three hit chemicals.
Results
These results suggest that Vallisneria spiralis L. extract contains various phytoconstituents, including carbohydrates, tannins, flavonoids, steroids, terpenoids, and glycosides. The distinctive functional groups of the extract were identified using a Fourier-Transform Infrared (FTIR) spectrophotometer. Nuclear Magnetic Resonance (NMR) spectroscopy was used to determine the structures of the isolated and purified extracts. The antifungal activity of Vallisneria spiralis L. extract against Candida albicans was found to be significantly stronger at a concentration of 200 mg/mL.
Conclusion
The findings of the study were obtained using in vitro and in silico methods to investigate the antifungal effect of Vallisneria spiralis L., which highlighted it as a potential natural antifungal treatment for Candida infection, which could be further investigated.
1. INTRODUCTION
Natural products and structural analogs have always played an important role in pharmacotherapy, particularly in the treatment of infections and malignancies [1, 2]. Natural drugs are categorized as authentic natural products, and drugs developed from natural products through semi-synthetic or synthetic processes are based on models of natural compounds [3, 4]. Phytochemicals, which are generally categorized as primary and secondary metabolites, are abundant in plants [5, 6]. Plant growth and development are driven by their main metabolites, which include proteins, lipids, and carbohydrates [7-9]. The three types of secondary metabolites, terpenoids, alkaloids, and phenolics, are primarily responsible for defensive mechanisms that enable plant survival [10, 11]. Physiologically active chemicals in plants must be isolated for their potential therapeutic applications, as well as their harmful effects. Secondary metabolite extraction, purification, and separation from plants are becoming increasingly common because of the pharmacological effects of phytochemicals [12]. A wide range of separation methods is employed in this process [13].
Vallisneria spiralis Linnaeus (Vallisneria spiralis L.), a dioecious perennial freshwater plant indigenous to southwest Asia, Middle East, northern Africa, and southern Europe (Fig. 1), is often referred to as tape grass, eelgrass, or eelweed [14]. Vallisneria spiralis L. belongs to the family of aquatic herbs known as Hydrocharitaceae, which is made up of many kinds of monocotyledonous, generally perennial plants. Vallisneria spiralis L. is a freshwater plant with fibrous roots, a short stem, and submerged, linear, tape-shaped leaves that are up to 10 mm broad and more than 100 cm long grouped in a basal rosette. The leaves have three–five parallel nerves and whole to finely serrated edges [15]. Owing to its thermophilic nature and preference for warmer climates, Vallisneria spiralis L. has expanded over tropical and subtropical regions as well as other regions outside its natural range, which have milder and more favorable climates. A perennial aquatic plant submerged in water, Vallisneria spiralis L., can have leaves as long as two meters or more, depending on the depth and flow of water [16]. A small vertical stem that produces rhizomes, which gives rise to new shoots, is where the cluster of leaves originates. The central longitudinal stripe is visible in the finely veined leaves. Each rosette base has fibrous, unbranched roots. The leaf edges range from being completely serrated to sharply rounded at the tips [17].
Candida is the predominant form of infection. A variety of factors can disrupt the normal balance of Candida species, which may cause an opportunistic infection. The proliferation of opportunistic Candida species can initiate a range of infections that include skin infections, urinary tract and vaginal infections, oral thrush, and bloodstream infections. Azole antifungals are the primary choice for the treatment or prevention of fungal infections. The long-term use of these medications (lasting weeks to months) is frequently accompanied by various undesirable effects [18, 19].
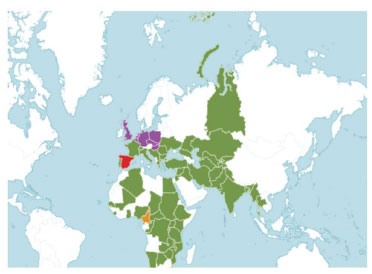
The map displays the global distribution of Vallisneria spiralis L., with native regions indicated in green, extinct regions in red, uncertain regions in orange, and introduced regions in purple [19].
The most commonly used antifungal medicines have several disadvantages in terms of toxicity, effectiveness, and cost. Additionally, their regular usage has contributed to the development of resistant strains. Natural products, whether in the form of standardized plant extracts or pure phytocompounds, provide many possibilities for novel therapeutic leads due to their unparalleled chemical variety. Plants contain many secondary metabolites that serve as significant reservoirs of chemical diversity. Therefore, they have the potential to be valuable sources of unexplored, powerful antibacterial and antifungal compounds. Preliminary qualitative screening of Vallisneria spiralis L. revealed the presence of many active compounds, including flavonoids, tannins, terpenoids, and glycosides, many of which possess antifungal properties. Studies have demonstrated that tannins can destroy bacterial membranes and deactivate their metabolism [20]. Flavonoids stimulate the production of extracellular soluble proteins and hinder metabolism, cell wall proteins, and DNA synthesis. Research has shown that flavonoid glycosides possess antibacterial properties. Their antibacterial effect is attributed to cell lysis and rupture of the cytoplasmic membrane by altering membrane permeability, resulting in the release of cellular components and ultimately causing cell death [21]. Terpenoids exhibit antifungal activity. The antifungal activity of terpenoid phenols is due to the inhibition of the target of the rapamycin (TOR) pathway and the effects of calcium stress. The TOR pathway is a signaling pathway that controls the metabolic status and cellular growth as a reaction to growth factors, nutrients, and other stimuli [22]. The incidence of fungal infections is on the rise, with over 300 million individuals worldwide afflicted by severe fungal diseases annually, as per global assessments [23].
Phytocompounds with antifungal properties can be explored as potential choices for the development of novel and enhanced alternative treatments for antifungal therapy. The current requirement for the effective treatment of fungal infections is the development of enhanced formulations, including plant phytocompounds. The inhibitory effect of Vallisneria spiralis L. phytocompounds on Candida albicans was demonstrated through qualitative screening of phytochemicals, anti-Candida assays, and in silico molecular docking techniques.
2. MATERIALS AND METHODS
2.1. Collection of Plant Materials
Vallisneria spiralis L. leaves were collected from the Kanyakumari in the Tamil Nadu state of India.
2.2. Extraction of Plant Material
Vallisneria spiralis L. leaves were initially pulverized into a fine powder using a blender. Subsequently, the powder was filtered through a 20 Mesh screen (850 µm) to guarantee uniformity. To create extracts with nonpolar and polar components, the dried leaf powder (400 g) was extracted using a Soxhlet apparatus (Fig. 2) for 8 h of refluxing chloroform and another 8 h of refluxing methanol [24]. The yields of the methanol and chloroform extracts were collected after evaporation to a constant weight.

The crude extracts are extracted using the soxhlet method.
2.3. Chromatographic Separation
The crude chloroform extract (27 g) from Vallisneria spiralis L. leaves was separated by flash column chromatography (Fig. 3) using a glass column measuring 90 cm L by 5 cm D and a silica gel stationary phase of 45–65 μm particle size and 63 Å porosity. The column was then packed with hexane. A step gradient of hexane/ethyl acetate (EtOAc) was used to elute the column (90% hexane, 5 L; 70% hexane, 4 L; 60% hexane, 3.2 L; 25% hexane, 2.8 L; 100% EtOAc, 2.5 L), and ethanol was used as a wash.
Thin-layer chromatography (TLC) was used to assess the obtained fractions [25]. TLC plates with a thickness of 200 μm were marked with a few drops of the concentrated fractions. After development in the proper solvent solution, the TLC plates were examined under a UV lamp [26]. After combining the fractions with similar TLCs, four significant superfractions were obtained. The vapor diffusion process was then used to recrystallize and purify these super fractions. Each superfraction was gently heated to dissolve in a small volume of EtOAc in a small vial. The vial was then placed in a bottle containing pentane and left undisturbed to form crystals. Superfraction one produced 450 mg of waxy, glossy, and colorless flakes. Superfractions three and four produced 2.16 g and 18.90 g of colorless crystals, respectively.
On a preparative TLC plate (20 × 20 cm, 250μm thick, silica gel supported on glass w/F-254), superfraction two was further fractionated. Superfraction two was administered in a line to the bottom of the plate using a capillary. The plate was developed to produce a subfraction when observed under UV light in a solvent chamber filled with hexane:EtOAc (85:15). Subsequently, a razor blade was used to scrape the appropriate bands from the glass. The compound was extracted from the adsorbent through dissolution in EtOAc and filtration through silica gel. Superfraction two produced 360 mg of colorless flakes.

Flash column chromatography.
2.4. Qualitative Analysis of Phytochemicals Extracted from Vallisneria Spiralis L.
The identification of plant chemicals with therapeutic potential depends heavily on phytochemistry, which is defined as the chemistry of plants and various plant components. Phytochemistry is regarded as an early branch of organic chemistry [27]. The Greek term “phyto” means “plant,” and phytochemicals are plant-based substances that are non-essential, have no nutritional benefit, and may even prevent illness [28]. It is possible to identify chemical groups or compounds in aqueous extracts from various plants using conventional analytical techniques and qualitative phytochemical investigations. The foundation of these qualitative tests is the appearance of colors or precipitates in response to the presence of these particular chemical components. The only thing that can be determined by all color reactions is whether particular chemical groups exist and not how much of them are present in various aqueous extracts [29].
2.4.1. Qualitative Screening of Carbohydrates
There are certain exceptions to the general rule that carbohydrates are neutral water-soluble chemical substances. Examples of these acids include pectic, gluconic, and alginic acids, which are found in living organisms.
2.4.2. Qualitative Screening of Tannins
Resorcinolic and pyrocatecholic aromatic rings are connected by a heterocyclic ring to form natural polyphenolic compounds known as tannins. Tannins are composed of flavan-3-ol repeating units [30].
2.4.3. Qualitative Screening of Flavonoids
Plants, fruits, and seeds contain large amounts of flavonoids, which are secondary metabolites that give them distinctive colors, flavors, and aromas. Flavonoids play a wide range of roles in plants, including controlling cell division, attracting pollinating insects, and providing defense against biotic and abiotic stressors [31].
2.4.4. Qualitative Screening of Steroids and Terpenoids
Terpenes and terpenoids are a broad class of secondary metabolites present in various plant groups. Two classes of isoprenoids, steroids and triterpenoids, are generated by the folding and cyclization of their common precursor, squalene [32].
2.4.5. Qualitative Screening of Glycosides
2.4.5.1. Test for Glycosides
Two milliliters of aqueous extract were mixed with a 0.5-milliliter solution of glacial acetic acid, two to three drops of ferric chloride were added, and 1 mL of concentrated sulfuric acid was added. The intense blue color that appeared at the interface of the two liquids confirmed the presence of cardiac glycosides.
2.4.6. Qualitative Screening of Saponins
One of the largest and most varied classes of naturally occurring plant compounds is saponins. Their ecological functions include protecting plants against diseases and herbivores and potentially acting as allelopathic agents in plant-to-plant competition [33].
2.4.7. Qualitative Screening of Alkaloids
Alkaloids are mostly produced from amino acids via biosynthesis, yielding a range of chemical structures that are primarily extracted from plants [34]. Alkaloids are a naturally occurring family of chemical molecules that include nitrogen and are widely distributed in plants [35].
2.4.8. Qualitative Screening of Proteins and Amino Acids
Every physiological activity that occurs in a live cell involves proteins. Proteins are insoluble in neutral ions, irreversibly coagulated upon heating, colloidal, and do not diffuse across the plasma membrane [36]. As amino acids include an amino and carboxylic acid moiety, they are amphoteric phytocompounds that are highly reactive and mostly soluble in water.
2.5. Test for Protein
2.5.1. Biuret Test
One milliliter of aqueous extract was mixed with two drops of 3% copper sulfate and a few drops of 10% sodium hydroxide; the production of a violet or red color indicated the presence of proteins [37].
2.5.2. Ninhydrin Test
Two drops of freshly prepared 0.2% ninhydrin solution were added to 1 mL of extract. If proteins are present, a purple color is produced [38].
2.7. Structural Elucidation of Phytochemicals Extracted from Vallisneria Spiralis L.
The distinctive functional groups in the extract were identified using a Fourier transform infrared (FTIR) spectrophotometer [39]. Five milligrams of the extract were dispersed in dry potassium bromide (KBr). To create a KBr thin disc, the mixture was thoroughly blended in a mortar and compressed for two minutes at a pressure of six bars. The disc was then placed in a diffuse-reflectance auxiliary sample cup. Infrared (IR) spectra were obtained using a Perkin Elmer 2000 IR spectrometer. To improve the signal-to-noise ratio, the sample was scanned 15 times from 400 to 4000cm-1. All sample runs were completed in a single run.
Nuclear Magnetic Resonance (NMR) spectroscopy was used to determine the structures of the isolated and purified extracts [40]. NMR spectra were collected using a Bruker Avance III 400 MHz spectrometer and processed using Topspin 3.1 software. We utilized deuterated methanol and chloroform. After dissolving each sample (7.5 mg in 750 μL of appropriate deuterated solvent), the temperature was measured at 25°C. A tetramethylsilane (TMS) reference standard was used to record the Proton (1H) spectra using a Varian NMR system 600 spectrometer operating at 600 MHz in DMSO-d6. The coupling constants (J) are reported in Hz, and the chemical shifts are recorded in ppm (δ).
2.8. In vitro Antifungal Assessment
2.8.1. Media Preparation
Antifungal screening on solid media involved the preparation of yeast peptone dextrose agar (YPDA) media. This media was created by combining 10 g yeast extract with 20 g peptone, glucose, and agar. The aforementioned materials were dissolved in 1 liter of distilled water and subsequently sterilized using an autoclave. The sterilized media was placed on autoclaved Petri plates and allowed to solidify at 25°C.
2.8.2. Culture Preparation
To create the culture, YPDA media was inoculated with Candida albicans ATCC code 10231. A small quantity of the culture was transferred to 5 mL of distilled water in a screw-capped test tube and agitated for 10 min to create a uniform suspension. The culture underwent overnight incubation at a temperature of 28°C.
2.8.3. Antifungal Screening
The susceptibility of the test compound to Candida albicans was assessed using disk diffusion [41, 42]. Three replicates of the assays were conducted, and the average results were recorded. The positive control employed in this study was fluconazole. A disk treated with dimethyl sulfoxide (DMSO) was used as the negative control. The minimum inhibitory concentrations (MICs) of the test and reference antifungal compounds were determined by the dilution method. Each disc was tested for antifungal activity at different concentrations (50, 75, 100, and 200 mg/mL). The discs were then incubated for 48 h at 28°C. Control experiments were conducted in an identical environment. The findings were recorded by quantifying the zone of inhibition in millimeters. To determine the MIC using the disc diffusion method, a zone of inhibition measuring 15 mm or more was deemed significant.
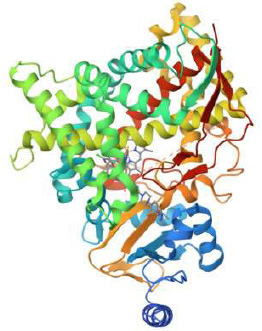
Three-dimensional (3D) structure of protein data bank (PDB) ID 5V5Z [49].
2.9. Molecular Docking Experiment
To investigate the inhibitory potential of our compounds against antifungal target proteins at the molecular level, a docking study was conducted to dock the three-dimensional structures of four extracted phytocompounds of Vallisneria spiralis L. into the receptor site of the target proteins using Autodock software. The three-dimensional (3D) structures of phytocompounds were generated using the Molinspiration Galaxy. The crystal structure of the target proteins (Fig. 4) was downloaded from the Protein Data Bank (PDB) using four codes. The PDB ID for the antifungal target was 5V5Z. Proteins and compounds were prepared according to a previously reported procedure [43]. After docking calculations, all conformations were analyzed based on their binding energies and interactions deep inside the active site of the proteins. Potential active site residues of the target proteins were determined through the utilization of the CASTp server. The grid box was set up on the co-crystallized ligand during the docking process, and the resulting measurements were documented in a config.txt file with the AutoDock Vina program [44]. The docked complexes were subjected to additional investi- gation and visualization for post-docking outcome analysis using GROMACS [45]. Biovia Discovery Studio was utilized to capture 2-D and 3-D illustrations [46]. The analysis of several molecular interactions implicated in the development of stable ligand-protein complexes was conducted using the Charmm GUI server [47, 48].
2.10. Prediction of Antifungal Activity and ADMET Properties
The PASS web application, i.e., Way2Drug, was used to predict antifungal activity [49-51]. An in silico approach was employed to evaluate the physicochemical, pharmaco- kinetic, and toxicological characteristics of phytochemicals extracted from Vallisneria spiralis. Screening of drug-like compounds was conducted using Lipinski's “rule of five” and a toxicity profile filter. Lipinski's drug-likeness characteristics were assessed using swissADME. ProTox was used to assess toxicity [52, 53].
3. RESULTS AND DISCUSSION
3.1. Qualitative Analysis of Phytochemicals
The presence of several phytoconstituents was revealed by a qualitative preliminary screening process conducted on different extracts of Vallisneria spiralis L. Table 1 lists the preliminary phytochemical analysis findings.
| Phytochemicals | Vallisneria Spiralis L. Extract |
|---|---|
| Carbohydrates | + |
| Tannins | + |
| Flavonoids | + |
| Steroids | + |
| Terpenoids | + |
| Glycosides | + |
| Saponins | - |
| Alkaloids | - |
| Proteins | - |
| Amino acids | - |
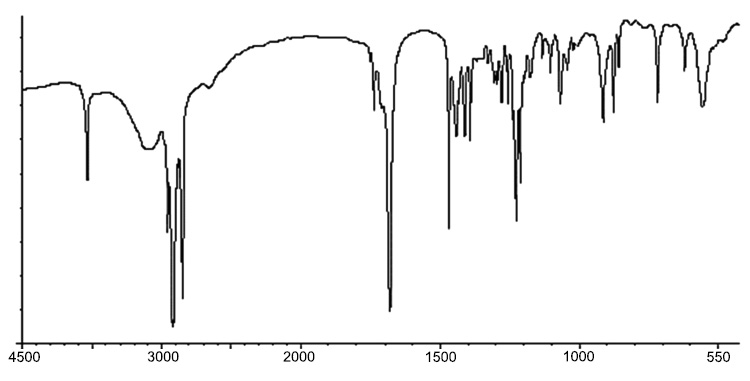
IR spectra of compound 1.
3.2. Spectroscopic Analysis
Using the FTIR spectrum, the peak value in the infrared radiation area was used to determine the functional groups of the active components. Figs. (5-10 and 11) correspond to IR spectra of Compound 1, Compound 2, Compound 3, and Compound 4, respectively. Different functional groups were found in the investigated compound when they were analyzed using infrared spectroscopy in a bandwidth ranging from 400 to 4000 cm-1. The phytochemical compounds were identified using NMR spectral data. The coupling constants (J) are reported in Hz, and chemical shifts are recorded in ppm (δ). Fig. (6, 8, 10, and 12) correspond to 1H NMR spectra of Compound 1, Compound 2, Compound 3, and Compound 4, respectively.
Compound 1: [4-(hydroxymethyl)-2,2,5-trimethylhexan- 3-yl] 2-methylpropanoate
IR: 3636, 2963, 2916, 1741, 1470, 1434, 1393 cm-1.
1H NMR: δ 0.82-0.94 (6H, 0.88 (d, J = 6.7 Hz), 0.88 (d, J = 6.7 Hz)), 0.94-1.05 (6H, 1.00 (d, J = 6.7 Hz), 1.00 (d, J = 6.7 Hz)), 1.06-1.18 (6H, 1.12 (d, J = 7.0 Hz), 1.12 (d, J = 7.0 Hz)), 1.35-1.53 (4H, 1.41 (o, J = 6.6 Hz), 1.41 (o, J = 6.6 Hz), 1.43 (tdd, J = 6.7, 6.6, 6.1 Hz), 1.45 (tdd, J = 7.1, 6.6, 6.1 Hz)), 2.42 (1H, sept, J = 7.0 Hz), 3.28-3.39 (2H, 3.33 (d, J = 6.7 Hz), 3.33 (d, J = 6.7 Hz)), 4.30-4.42 (2H, 4.36 (d, J = 7.1 Hz), 4.36 (d, J = 7.1 Hz)).
Compound 2: (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate
IR: 3512, 2963, 2934, 1736, 1486, 1460, 886, 798 cm-1.
1H NMR: δ 0.87 (9H, s), 0.96-1.17 (9H, 1.02 (d, J = 6.8 Hz), 1.11 (d, J = 7.0 Hz), 1.11 (d, J = 7.0 Hz)), 2.10 (1H, hd, J = 6.8, 2.7 Hz), 2.40 (1H, sept, J = 7.0 Hz), 3.34 (1H, d, J = 2.7 Hz), 4.29-4.40 (2H, 4.34 (d, J = 6.8 Hz), 4.34 (d, J = 6.8 Hz)).
Compound 3: 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0] heptan-1-yl)but-3-en-2-one
IR: 3258, 2979, 2681, 2660, 1698, 1475, 1324, 1182, 911 cm-1.
1H NMR: δ 0.99-1.09 (6H, 1.04 (s), 1.04 (s)), 1.26-1.46 (4H, 1.34 (ddd, J = 13.3, 4.1, 1.9 Hz), 1.41 (s)), 1.55-1.92 (5H, 1.63 (ddddd, J = 12.9, 6.3, 4.1, 1.9, 1.4 Hz), 1.69 (ddd, J = 13.3, 10.0, 1.9 Hz), 1.75 (ddd, J = 13.1, 8.2, 6.3 Hz), 1.82 (ddddd, J = 12.9, 10.0, 8.2, 6.2, 1.9 Hz), 1.83 (ddd, J = 13.1, 6.2, 1.4 Hz)), 2.10-2.24 (5H, 2.15 (s), 2.18 (t, J = 7.3 Hz), 2.18 (t, J = 7.3 Hz)), 2.46-2.58 (2H, 2.52 (t, J = 7.3 Hz), 2.52 (t, J = 7.3 Hz)).
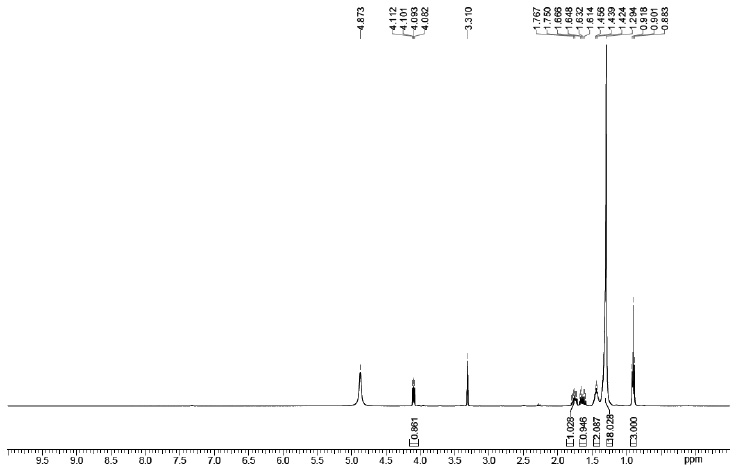
1H NMR spectra of compound 1.
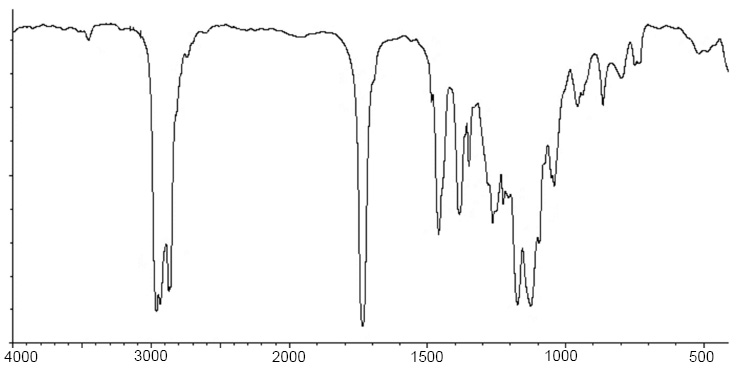
IR spectra of compound 2.
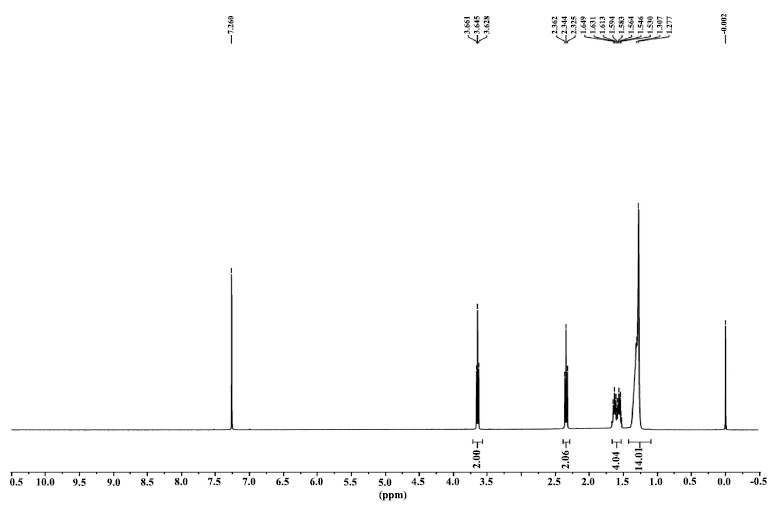
1H NMR spectra of compound 2.
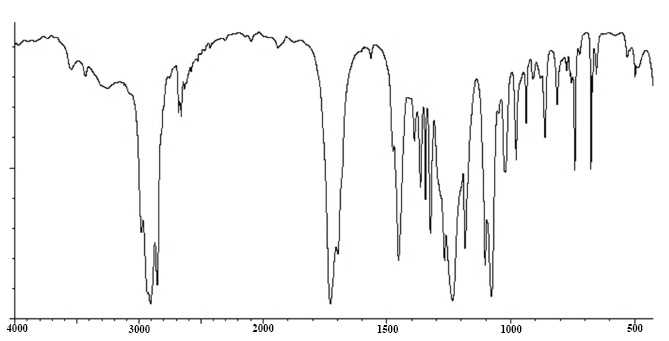
IR spectra of compound 3.
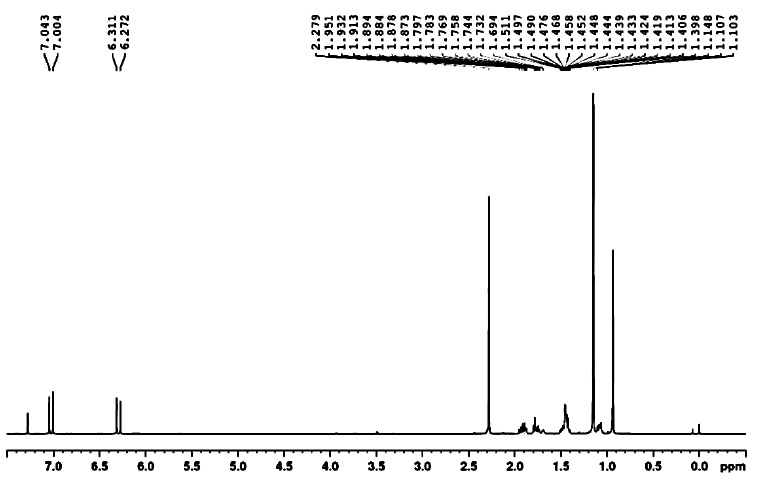
1H NMR spectra of compound 3.
Compound 4: 2,2,6-trimethylcyclohexane-1,4-dione
IR: 2931, 2872, 1467, 1370, 1309, 1281, 1154, 1119, 985 cm-1.
1H NMR: δ 1.07-1.25 (9H, 1.12 (s), 1.12 (s), 1.19 (d, J = 6.6 Hz)), 2.34-2.67 (4H, 2.41 (d, J = 15.9 Hz), 2.50 (d, J = 15.9 Hz), 2.58 (dd, J = 15.7, 4.0 Hz), 2.59 (dd, J = 15.7, 10.0 Hz)), 2.92 (1H, dqd, J = 10.0, 6.6, 4.0 Hz).
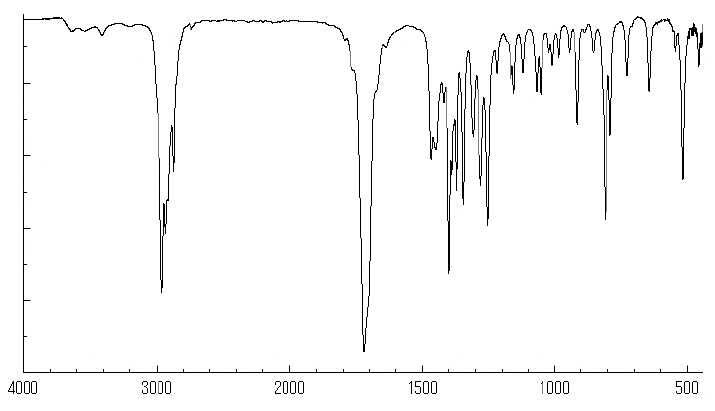
IR spectra of compound 4.
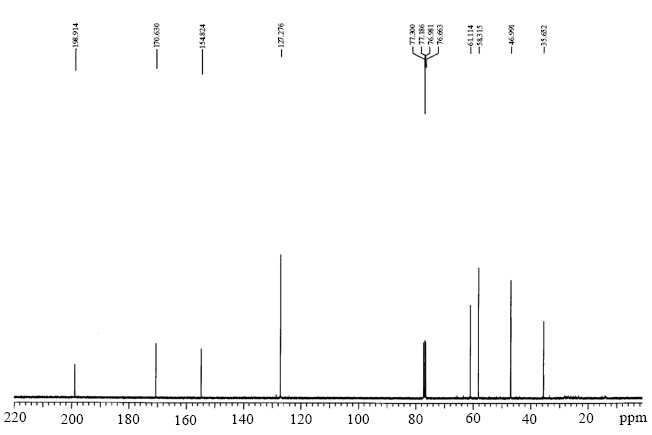
1H NMR spectra of compound 4.
3.3. In Vitro Antifungal Activity
This study aimed to evaluate the in-vitro antifungal properties of methanolic extracts derived from Vallisneria spiralis L. against Candida albicans. The extracts were evaluated at concentrations of 200 mg/mL, 100 mg/mL, 75 mg/mL, and 50 mg/mL. The findings from the assessment of antifungal activity indicated that Vallisneria spiralis L. has a significant inhibitory effect against Candida albicans (Table 2). The antifungal activity of Vallisneria spiralis L. extracts against Candida albicans was found to be significantly stronger at a concentration of 200 mg/mL, as evidenced by the presence of inhibition zones of 18.5 ± 0.5 mm. The acquired data were recorded in Microsoft Excel and subsequently analyzed using data analysis software, namely STATA version 14. To examine the variability in the concentrations of Vallisneria spiralis L. extract, a one-way analysis of variance (ANOVA) was conducted. A post-hoc Tukey test was employed to assess the relationship between the zone of inhibition across different concentrations and the negative control. Differences were considered statistically significant if the p-value was less than 0.05.
3.4. Prediction of ADMET Properties
The detailed in silico ADMET analysis of the Vallisneria spiralis L. phytocompounds is presented in Tables 3 and 4. Based on the toxicological investigation of Vallisneria spiralis L. phytocompounds, it can be concluded that the phytocompounds did not show any signs of toxicological concerns.
| Concentration | Inhibition Zone (mm) |
|---|---|
| Vallisneria spiralis 50 mg/mL | 10.8 ± 0.5a |
| Vallisneria spiralis 75 mg/mL | 12.6 ± 0.5a |
| Vallisneria spiralis 100 mg/mL | 14.4 ± 0.5a,b |
| Vallisneria spiralis 200 mg/mL | 18.5 ± 0.5a,c |
| Fluconazole 25 mg/mL | 22.1 ± 0.5 |
| DMSO 5% (negative control) | — |
a the observed difference was significant (p < 0.05) compared with the negative control;
b there was a statistically significant difference (p < 0.05) among the concentrations of 100 , 50, and 75 mg/mL.
c the observed difference was significant (p < 0.05) between the concentrations of 200, 50, 75, and 100 mg/mL.
| - | SwissADME | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug-likeness Rules | |||||||||||
| - | Lipinski’s rule of 5 | - | |||||||||
| Phytochemicals | Water Solubility | GI Absorption | LOGP | Hydrogen bond donor |
Hydrogen bond acceptor |
TPSA (Å2) | MW | Ghose | Veber | Egan | Muegge |
| [4-(hydroxymethyl)-2,2,5-trimethylhexan-3-yl] 2-methylpropanoate | Soluble | High | 3.21 | 1 | 3 | 46.53 | 244.37 | Yes | Yes | Yes | Yes |
| (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate | Soluble | High | 2.84 | 1 | 3 | 46.53 | 216.32 | Yes | Yes | Yes | Yes |
| 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl)but-3-en-2-one | Soluble | High | 2.83 | 0 | 2 | 29.60 | 208.30 | Yes | Yes | Yes | Yes |
| 2,2,6-trimethylcyclohexane-1,4-dione | Soluble | High | 1.63 | 0 | 2 | 34.14 | 154.21 | No | Yes | Yes | No |
| - | Protox II | ADMET SAR | ||||
|---|---|---|---|---|---|---|
| Phytochemicals | LD50 mg/kg | Hepatotoxicity | Mutagenicity | AMES Toxicity | Tetrahymena Pyriformis Toxicity pIGC50 ug/L |
Rat Acute Toxicity LD50 mol/kg |
| [4-(hydroxymethyl)-2,2,5-trimethylhexan-3-yl] 2-methylpropanoate | 8350 | No | No | No | -0.2728 | 1.7762 |
| (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate | 3200 | No | No | No | -0.2129 | 1.7990 |
| 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl)but-3-en-2-one | 2000 | No | No | No | 0.5470 | 1.7880 |
| 2,2,6-trimethylcyclohexane-1,4-dione | 2400 | No | No | No | 0.5230 | 2.3921 |
3.5. In Silico Antifungal Activity
The antifungal potential of the three best compounds, i.e., Compound 1: [4-(hydroxymethyl)-2,2,5-trimethyl- hexan-3-yl] 2-methylpropanoate, Compound 2: (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate, and Compound 3: 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan- 1-yl)but-3-en-2-one, extracted phytocompounds of Vallisneria spiralis L., was explored by docking into the active site of the antifungal target (PDB ID = 5V5Z). The inhibitory effects of Vallisneria spiralis L. phytocompounds on Candida albicans are presented in Table 5. The Pa values of Vallisneria spiralis L. phytocompounds were higher than those of Pi, indicating a likelihood of biological activity rather than inactivity in inhibiting Candida albicans. This demonstrates the higher likelihood of these specific chemicals inhibiting the growth of C. albicans. Phytocompounds from Vallisneria spiralis L. indicate a greater likelihood of exhibiting antifungal action.
| Compound No. | Chemical Name | Pa | Pi |
|---|---|---|---|
| 1 | [4-(hydroxymethyl)-2,2,5-trimethylhexan-3-yl] 2-methylpropanoate | 0.546 | 0.024 |
| 2 | (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate | 0.446 | 0.040 |
| 3 | 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl)but-3-en-2-one | 0.285 | 0.087 |
Compound 1 showed conventional hydrogen bonding interactions with key amino acid Gln67 and pi-alkyl interactions with Tyr69 and His373 of the target protein 5V5Z (Fig. 13). Compound 2 showed hydrogen bonding interactions with important residues of the active sites His373, Tyr69, Met372, and pi-alkyl interactions with His410, His373, and Met374 (Fig. 14). Compound 3 interacted through conventional hydrogen bonding interactions with key residues Gln67 and Tyr505 and showed pi-alkyl interactions with His373, Pro503, Val510, and Tyr69 (Fig. 15). The binding energies for the simulated protein (5V5Z) and ligands are listed in Table 6.
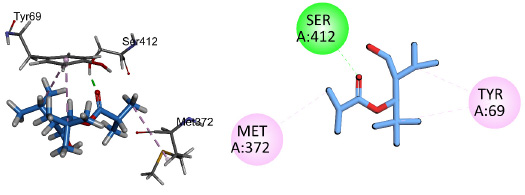
2D and 3D interactions of compound 1 with antifungal target protein 5V5Z.
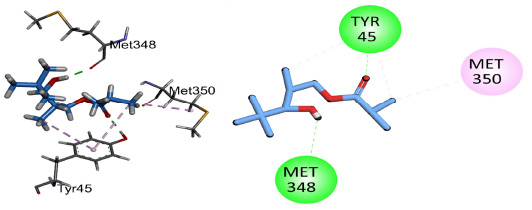
2D and 3D interactions of compound 2 with antifungal target protein 5V5Z.
| Compound No. | Chemical Name | Docking Compound Code | Molecular Formula | PDB ID | Binding Energy | Amino Acid Involved in Hydrogen Bonding |
|---|---|---|---|---|---|---|
| 1 | [4-(hydroxymethyl)-2,2,5-trimethylhexan-3-yl] 2-methylpropanoate | 5V5Z1 | C14H28O3 | 5V5Z | -3.90 | Gln67 |
| 2 | (3-hydroxy-2,4,4-trimethylpentyl) 2-methylpropanoate | 5V5Z2 | C12H24O3 | 5V5Z | -4.56 | His373, Tyr69, Met372 |
| 3 | 4-(2,2,6-trimethyl-7-oxabicyclo[4.1.0]heptan-1-yl)but-3-en-2-one | 5V5Z3 | C13H20O2 | 5V5Z | -5.51 | Gln67, Tyr505 |
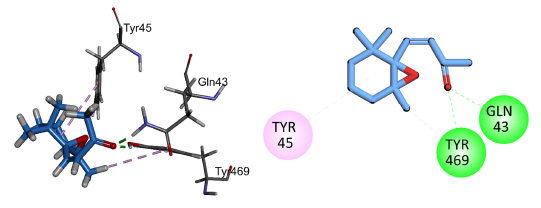
2D and 3D interactions of compound 3 with antifungal target protein 5V5Z.
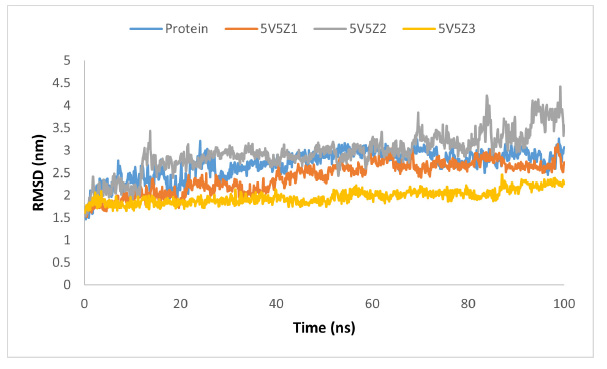
RMSD graph of protein and protein-ligand complex.
The molecular-dynamic simulations were used to assess the inhibitory capability and influence of the best three phytocompounds from Vallisneria spiralis L. on the structural and dynamic features of the target protein. To assess the structural and conformational changes in the protein before and after complex formation, essential measures, such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bonding interaction, radius of gyration, and PCA analysis were used. RMSD was used to assess the protein's structural and configuration changes during the simulation (100 ns). The protein (blue) showed almost steady behavior between 1.5 and 3 nm; however, some more frequent fluctuations in RMSD were observed in the range of 0 to 30 ns. after 30 ns, showing protein (5V5Z) stability with little deviation. The protein and compound-1 complex (5V5Z1) (orange color) showed average deviation and RMSD between 1.5 and 3.0 nm with moderate steady behavior. In contrast, the protein and ligand-2 complex (5V5Z2) (gray color) exhibited the highest deviation with an RMSD range of 1.5–4.5 nm. The highest deviation was observed at 10–20 ns and 70–100 ns, indicating conformational changes in the complex during the simulation. Compound-3 and the protein complex (5V5Z3) (yellow color) showed the highest stability with a minimum RMSD in the range of 1.5–2.5 nm (Fig. 16).
RMSF analysis was performed to assess the variability of the residues and other protein components during the simulation (Fig. 17). The RMSF graph indicates that proteins and protein-ligand complexes fluctuate in the same way but with varying intensities and few exceptions. Proteins showed higher volatility than complexes. 5V5Z1 and 5V5Z3 exhibited less volatility than 5V5Z2. The residues, 25-36, 229-242, 348-361, and 425-453 in sequence, exhibited the most variability in the protein; however, the same areas showed minimal or very little fluctuation in the complexes.
Hydrogen bonding interactions were investigated to confirm the docking results, explain the inhibitory potential, and specify the number of hydrogen bonds during the molecular dynamic (MD) simulation (Fig. 18). The 5V5Z1 data demonstrated that ligand-1 residues up to 670 exhibited significant hydrogen bonding interactions, with a maximum of four hydrogen bonds and an average of two hydrogen bonding interactions. However, residues after 670 exhibited little or no interaction with the ligand. The ligand-2 in compound 5V5Z2 exhibited the same interactions as 5V5Z1 with a few additions. Throughout the MD simulations, ligand-3 consistently exhibited a single hydrogen bonding interaction, resulting in a stable behavior.
Protein conformational alterations and conformational stability were examined using principal component analysis (PCA). According to PCA, proteins have a wide conformational space prior to ligand inhibition. However, proteins have less conformational space after complexation with ligands. All complexes had only the following conformational states: 5V5Z1, 5V5Z2, and 5V5Z3. In contrast to 5V5Z1 and 5V5Z2, which exhibited very little reduction, the conformational space in 5V5Z3 was greatly reduced. A 2D PCA plot is shown in Fig. (19).
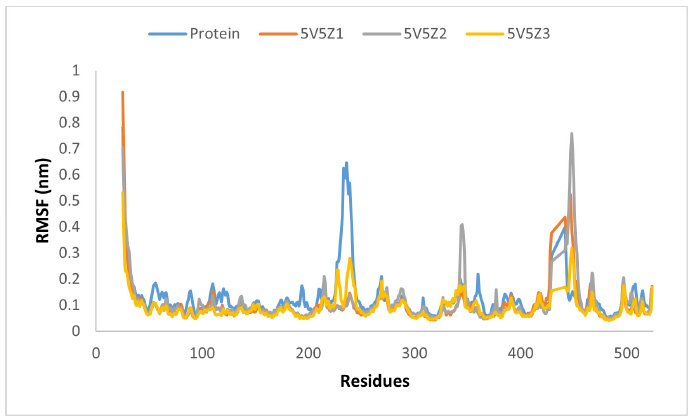
The RMSF graph of protein and our ligands.
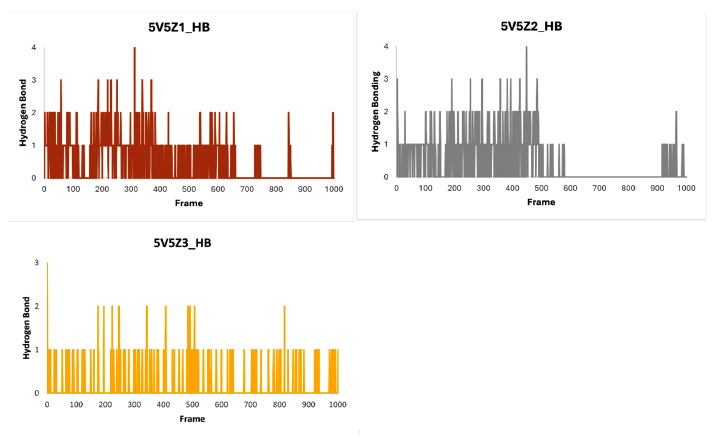
Hydrogen bonding histograms of ligands 5V5Z1, 5V5Z 2 and 5V5Z 3 with target protein 5V5Z.
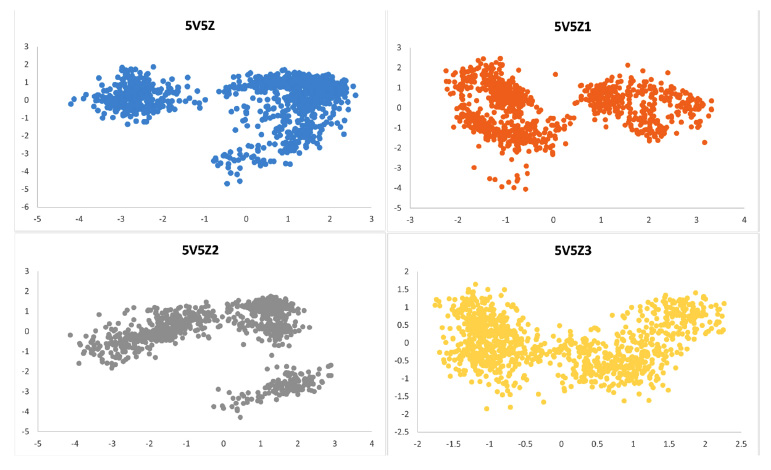
Principal component analysis (PCA) of protein 5V5Z, 5V5Z1, 5V5Z2, and 5V5Z3.
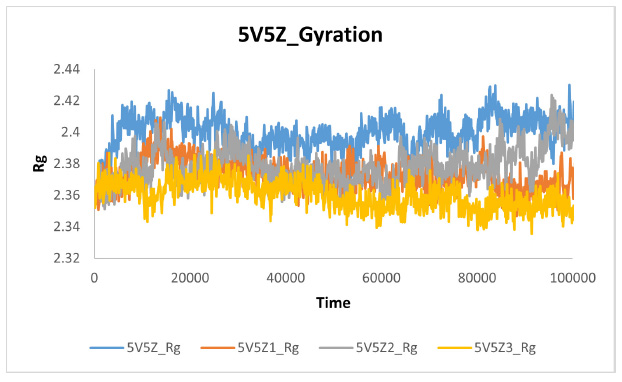
The gyration plot of protein and ligands.
Protein compactness was investigated during MD simulations using a gyration plot. When compound-1, compound-2, and compound-3 formed a complex with the target protein 5V5Z, compactness increased (the size decreased), and fluctuations decreased. These findings demonstrated that, in comparison to 5V5Z1 and 5V5Z2, 5V5Z3 was more compact and stable in size. Fig. (20) illustrates the comparative gyration plot.
CONCLUSION
The aim of this study was to conduct a preliminary phytochemical analysis and assessment of Vallisneria spiralis L. extract. An attempt was undertaken to analyze isolated phytoconstituent derived from Vallisneria spiralis L. Phytochemical screening of Vallisneria spiralis L. yielded four compounds. A qualitative investigation was performed to characterize the extracted and purified phytoconstituents. The recovered phytoconstituents were characterized using advanced spectral methods, such as FTIR and 1H NMR. Four phytochemicals of Vallisneria spiralis L. were discovered and reported for the first time. In-vitro and in-silico antifungal characteristics demonstrated that Vallisneria spiralis L. has the potential to be an antifungal candidate.
AUTHORS’ CONTRIBUTION
R.W. and S.K.P. contributed to the conceptualization of the study and proposed the methodology. R.W. carried out the formal analysis and was involved in the investigation, data curation, and writing the original draft preparation. S.K.P. took part in writing the review, editing, supervision, and project administration. All authors have read and agreed to the published version of the manuscript.
LIST OF ABBREVIATIONS
| TOR | = Target of the Rapamycin |
| NMR | = Nuclear Magnetic Resonance |
| FTIR | = Fourier-Transform Infrared |
| IR | = Infrared |
| YPDA | = Yeast Peptone Dextrose Agar |


