All published articles of this journal are available on ScienceDirect.
Analysis of PTGS1 and PEAR1 Polymorphisms for Aspirin Resistance Prediction in Patients with Cerebral Infarction
Abstract
Introduction
This study aimed to evaluate the predictive potential of prostaglandin-endoperoxide synthase 1 (PTGS1) and platelet endothelial aggregation receptor-1 (PEAR1) gene polymorphisms, combined with biochemical and demographic factors, for aspirin resistance (AR) and recurrence of cerebral infarction (CI) in Chinese patients.
Methods
A retrospective study was conducted on 280 CI patients admitted to the Department of Neurology, Affiliated Hospital of Beihua University, between January and December 2021. Based on telephone follow-up, patients were divided into recurrent (n = 97) and non-recurrent (n = 183) groups. Clinical characteristics, medical history, and laboratory indicators were collected. Genotyping for PEAR1 and PTGS1 was performed. Logistic regression identified independent predictors, and receiver operating characteristic (ROC) curves evaluated predictive performance.
Results
Univariate analysis showed significant differences in PEAR1 and PTGS1 genotypes, hypertension, and homocysteine (HCY) levels between groups (P < 0.05). Multivariate analysis showed PEAR1, PTGS1, and HCY as independent predictors of recurrence (p < 0.05). ROC analysis revealed AUC values as follows: PEAR1 = 0.529, PTGS1 = 0.642, HCY = 0.696, and combined model = 0.721.
Discussion
PEAR1 may influence platelet activation, while PTGS1 can alter COX-1 function, reducing aspirin’s antiplatelet efficacy. Elevated HCY promotes endothelial dysfunction and thrombosis. The combined model offered superior predictive value, supporting its potential clinical utility in guiding antiplatelet therapy for individualized risk stratification. However, this single-center study with a limited sample size requires validation in larger multicenter cohorts.
Conclusion
PEAR1 and PTGS1 genes integrated with HCY can effectively predict the risk of recurrence in patients with cerebral infarction.
1. INTRODUCTION
Ischemic stroke, a leading subtype of cerebral infarction, remains a major cause of disability and mortality worldwide [1]. With the intensification of the aging trend of the population in our country and the prevalence of unhealthy lifestyles, the incidence of stroke has been increasing year by year [2]. Early deterioration of neurological function and recurrence in patients are more common, and they seriously affect people's quality of life [3]. Despite advances in acute stroke management, secondary prevention strategies are crucial to reducing the risk of recurrence. Among these, antiplatelet therapy, particularly with aspirin [4], plays a pivotal role. However, a subset of patients exhibits aspirin resistance [5], a condition characterized by the failure of aspirin to adequately inhibit platelet aggregation, thereby increasing the likelihood of recurrent vascular events.
The mechanisms underlying aspirin resistance are complex and multifactorial, involving biochemical and clinical factors [6]. At a pharmacological level, aspirin prevents stroke primarily through irreversible, dose-dependent inhibition of the cyclooxygenase (COX) enzyme, with a particular affinity for the COX-1 isoform in platelets [7]. COX-1 catalyzes the conversion of arachidonic acid to prostaglandin H2, the precursor of thromboxane A2, a potent promoter of platelet aggregation and vasoconstriction. By acetylating a serine residue in the COX-1 active site, aspirin suppresses TXA2 synthesis, thereby reducing platelet activation and aggregation.
Recent studies have highlighted the significance of gene polymorphisms in modulating individual responses to antiplatelet agents. Notably, gene polymorphisms involving PEAR1 and PTGS1, which are directly involved in platelet activation and thromboxane A2 synthesis, have been implicated in AR [8]. Consequently, these gene polymorphisms may lead to variations in platelet activity among patients with cerebrovascular disease [9].
Additionally, several biochemical markers, such as elevated HCY, and comorbid conditions, like hypertension and diabetes, have also been associated with increased risk of recurrent stroke and poor antiplatelet response [10]. Integrating genetic and biochemical indicators could therefore enhance the prediction of AR and recurrence in patients with cerebral infarction. The current study has addressed a knowledge gap by developing and evaluating a model, thereby providing a novel tool for early identification of at-risk patients.
This study aimed to investigate the predictive value of PEAR1 and PTGS1 gene polymorphisms, in combination with biochemical and demographic indicators, for aspirin resistance and recurrence in patients with cerebral infarction. By identifying patients at higher risk for AR, our findings may inform more personalized and effective secondary prevention strategies.
2. METHODS
2.1. Ethics Approval
This study has been approved by the ethics committee of Beihua University Affiliated Hospital (reference no.: 2024005), and all patients included have signed the informed consent. The Helsinki Declaration has been followed for involving human subjects in the study.
2.2. Study Design and Participants’ Recruitment
A total of 280 patients diagnosed with cerebral infarction who were admitted to the Department of Neurology, Beihua University Affiliated Hospital, from January 2021 to December 2021, were enrolled in this study. The follow-up was conducted through scheduled telephone interviews with patients or their immediate family members, and recurrence of cerebral infarction was verified by newly reported neurological symptoms, which were cross-checked against hospital admission records and available diagnostic imaging results. Patients were categorized into two groups: a recurrent group (n = 97) and a non-recurrent group (n = 183). Inclusion criteria were as follows: (1) diagnosed with cerebral infarction confirmed by head CT or MRI; (2) absence of other neurological diseases or craniocerebral diseases; and (3) administration of antiplatelet therapy with aspirin alone (100 mg/day) or dual antiplatelet therapy with aspirin (100 mg/day) and clopidogrel (75 mg/day) after admission. Exclusion criteria were as follows: (1) allergy to aspirin or a history of asthma; (2) severe liver diseases or coagulation disorders; (3) platelet count < 100×109/L or > 300×109/L; (4) bleeding tendency or presence of gastric ulcer; (5) cerebral infarction primarily caused by cardiogenic embolism; (6) history of trauma, cerebral hemorrhage, or surgery within the past two weeks; (7) history of hemorrhagic disorders; (8) lost to follow-up; and (9) poor medication compliance.
2.3. Sociodemographic Factors, Clinical Information, and Blood Sampling
Demographic and clinical characteristics, including age, sex, and history of hypertension, diabetes mellitus, coronary artery disease, smoking, and alcohol consumption, were retrospectively collected. Laboratory parameters obtained at the time of hospital admission, including hemoglobin concentration, triglyceride levels, total cholesterol levels, serum uric acid, and plasma HCY levels, were recorded.
2.4. Genotyping
SNP genotyping was performed using a fluorescence-based real-time PCR system. After instrument self-check and preheating, the analysis software was launched, and the appropriate gene panel was selected. Sample identifiers and corresponding genotype assays were entered, and settings were saved. Fluorescently labeled probes were hybridized to target SNP alleles, and the fluorescence detector monitored signal changes in real time to distinguish alleles based on emission patterns. Genotypes were automatically assigned by the software and stored for analysis. Quality control procedures included duplicate testing of 10% of randomly selected samples, which yielded a concordance rate of 100%. The overall genotyping error rate was <1%, confirming high assay reliability.
2.5. Statistical Analysis
Measurement data conforming to a normal distribution have been presented as mean ± standard deviation (x̄ ± s), and comparisons between groups were performed using the independent samples t-test. Categorical data have been expressed as frequencies and percentages, and comparisons between groups were conducted using the chi-square (χ2) test. First, a univariate analysis of all relevant variables was performed. Then, the variables with statistical significance in the univariate analysis were included in the multiple logistic regression model. Finally, independent predictors were identified, and a combined prediction model was constructed. The diagnostic performance of the single and combined predictive models based on PEAR1 and PTGS1 genotypes for evaluating AR in patients with cerebral infarction was assessed using ROC analysis, and the AUC was compared. A p-value < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS software, version 31.0 (IBM Corp., Armonk, NY, USA).
3. RESULTS
3.1. Characteristics of the Patients
A total of 280 patients were enrolled in this study, comprising 97 individuals (34.6%) in the recurrence group and 183 individuals (65.4%) in the non-recurrence group. Of the total cohort, 210 patients (75.0%) were male, and 70 patients (25.0%) were female. The mean age of all participants was 63.21 ± 10.16 years. The recurrence group had a mean age of 64.27 ± 10.03 years, while the non-recurrence group had a mean age of 62.11 ± 10.22 years.
3.2. Univariate Logistic Regression Analysis
Table 1 presents the univariate analysis comparing demographic, clinical, biochemical, and genetic variables between cerebral infarction patients with recurrence (n=97) and without recurrence (n=183). Significant differences were observed for hypertension (p = 0.024), HCY levels (p = 0.001), and the distribution of PEAR1 (p = 0.010) and PTGS1 (p < 0.001) genotypes.
| Characteristics | Non-recurrence Group (n=183) | Recurrence Group (n=97) | p-value |
|---|---|---|---|
| Age (mean ± SD) | 62.11±10.22 | 64.27±10.03 | 0.092 |
| #Gender, n (%) | 143.00 (78.14) | 67.00 (69.07) | 0.095 |
| Previous history, n (%) |
- | - | - |
| Hypertension | 107.00 (58.47) | 70.00 (72.16) | 0.024 |
| Diabetes | 57.00 (31.15) | 37.00 (38.14) | 0,238 |
| Coronary heart disease | 53.00 (28.96) | 36.00 (37.11) | 0.163 |
| Smoking, n (%) | 92.00 (50.27) | 51.00 (52.58) | 0.714 |
| Alcohol drinking, n (%) |
80.00 (43.72) | 42.00 (43.30) | 0.947 |
| Biochemical parameters (mean ± SD) | - | - | - |
| Hemoglobin | 145.10 ± 17.10 | 146.60 ± 18.14 | 0.493 |
| Triglycerides | 1.69 ± 1.11 | 1.92 ± 1.39 | 0.128 |
| Total cholesterol | 4.35 ± 1.04 | 4.40 ± 1.08 | 0.314 |
| HCY | 15.12 ± 12.95 | 20.87 ± 14.35 | 0.001 |
| Uric acid | 332.32 ± 80.11 | 347.20 ± 97.73 | 0.172 |
| Gene polymorphisms | - | - | - |
| PEAR1, n (%) | - | - | - |
| AG | 71.00 (38.80) | 71.00 (73.20) | - |
| GG | 97.00 (53.01) | 11.00(11.34) | 0.010 |
| AA | 15.00 (8.20) | 15.00 (15.46) | - |
| PTGS1, n (%) | - | - | - |
| AA | 121.00 (66.12) | 62.00 (63.92) | - |
| AG | 50.00 (27.32) | 29.00 (29.90) | < 0.001 |
| GG | 12.00 (6.56) | 6.00 (6.19) | - |
3.3. Multivariate Logistic Regression Analysis
Multivariate logistic regression analysis showed elevated HCY (OR = 1.031, 95% CI: 1.011–1.052, p = 0.002), PEAR1 polymorphism (OR = 0.144, 95% CI: 0.047–0.437, p = 0.001), and PTGS1 polymorphism (OR = 0.196, 95% CI: 0.072–0.532, p = 0.001) to be independent predictors. Hypertension and previous history were not statistically significant in the adjusted model (Table 2).
| Characteristics | B | SE | p-value | OR | 95%CI |
|---|---|---|---|---|---|
| Previous history | - | - | - | - | - |
| Hypertension | - 0.100 | 0.330 | 0.762 | 0.905 | 0.474-1.729 |
| Biochemical parameters | - | - | - | - | - |
| HCY | 0.031 | 0.010 | 0.002 | 1.031 | 1.011-1.052 |
| Gene polymorphisms PEAR1 |
-1.940 | 0.568 | 0.001 | 0.144 | 0.047-0.437 |
| PTGS1 | -1.629 | 0.509 | 0.001 | 0.196 | 0.072-0.532 |
| Constant | 1.471 | 0.701 | 0.036 | 4.354 | - |
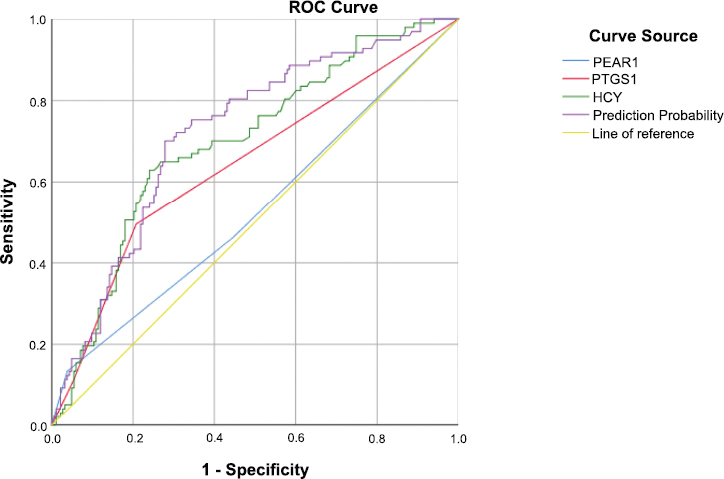
ROC analysis of genetic and biochemical predictors of cerebral infarction recurrence.
3.4. Model Evaluation and ROC Curve Analysis
ROC curves showing the predictive performance of PEAR1, PTGS1, and HCY levels, as well as their combined prediction probability, for recurrence of cerebral infarction (Fig. 1). The AUC was highest for the combined model, indicating improved discriminative ability compared to individual predictors. The yellow diagonal line represents the reference line (AUC = 0.5).
ROC curve analysis (Table 3) demonstrated the predictive performance of the PEAR1 gene alone to be limited (AUC = 0.529, 95% CI: 0.456–0.602, p = 0.424), indicating poor discriminatory ability. The PTGS1 gene showed improved discrimination (AUC = 0.642, 95% CI: 0.572–0.712, p < 0.001), corresponding to fair predictive accuracy. Plasma HCY yielded a higher AUC of 0.696 (95% CI: 0.632-0.761, p < 0.001), reflecting moderate predictive ability. The combined prediction model incorporating genetic and biochemical markers achieved the highest discriminative performance (AUC = 0.721, 95% CI: 0.659-0.783, p < 0.001), suggesting moderate overall predictive capability.
| Characteristics | AUC | SE | p-value | 95%CI |
|---|---|---|---|---|
| Gene polymorphisms | - | - | - | - |
| PEAR1 | 0.529 | 0.037 | 0.424 | 0.456-0.602 |
| PTGS1 | 0.642 | 0.036 | < 0.001 | 0.572-0.712 |
| Biochemical parameters | - | - | - | - |
| HCY | 0.696 | 0.033 | < 0.001 | 0.632-0.761 |
| Prediction probability | 0.721 | 0.032 | < 0.001 | 0.659-0.783 |
4. DISCUSSION
In this study of 280 patients with cerebral infarction, multivariate logistic regression identified PEAR1, PTGS1, and HCY as independent predictors of cerebral infarction recurrence. The combined predictive model incorporating these three markers achieved an AUC of 0.721, outperforming any single marker alone and suggesting moderate discriminatory ability, suggesting that the model can reliably distinguish between patients at higher versus lower risk of recurrence in approximately 72% of cases.
Our findings have been found to be consistent with previous studies [11-13], linking elevated HCY levels to increased risk of ischemic cerebrovascular events, supporting the role of HCY as a modifiable biochemical risk factor. Elevated plasma HCY levels can exacerbate vascular injury through endothelial dysfunction, oxidative stress, and promotion of a prothrombotic state [14, 15].
Similarly, PEAR1 polymorphisms have been associated with altered platelet aggregation and recurrent ischemic stroke [16-18], while PTGS1 polymorphism may impair aspirin’s antiplatelet effect by modifying prostaglandin metabolism [19, 20]. Gene polymorphisms in PEAR1 and PTGS1 may alter platelet receptor signaling and cyclooxygenase activity [21, 22], respectively, thereby reducing aspirin’s ability to inhibit platelet aggregation.
The coexistence of these genetic and biochemical risk factors likely produces additive or synergistic effects, resulting in a higher likelihood of aspirin resistance and increased risk of recurrent cerebrovascular events (Fig. 2). Clinically, this combined genetic-biochemical model offers a more comprehensive risk stratification approach. Routine genetic testing for PEAR1 and PTGS1 is not yet standard practice in China or elsewhere; however, the current results suggest that it could be valuable for patients at high risk of recurrence, particularly when combined with HCY measurement. Identifying high-risk individuals before treatment could guide tailored antiplatelet strategies, such as alternative antiplatelet agents, dose adjustments, or concurrent therapies targeting elevated HCY levels, including folate and B-vitamin supplementation [23, 24].
4.1. Limitations of the Study
There are certain limitations that need to be considered. Firstly, this study enrolled patients with cerebral infarction from Jilin Province, Northeast China. Accordingly, the findings have predominantly reflected the genetic and environmental characteristics of this regional population and may not be generalizable to diverse ethnic groups within China, or to global cohorts. Secondly, a platelet function test was not performed in this study to define the incidence of aspirin resistance. Similarly, this study only investigated two genes related to aspirin resistance. Finally, in this study, there were relatively few recurrence observation indicators, which could not prove a strong correlation between cerebral infarction recurrence and secondary endpoint events, as well as aspirin resistance.
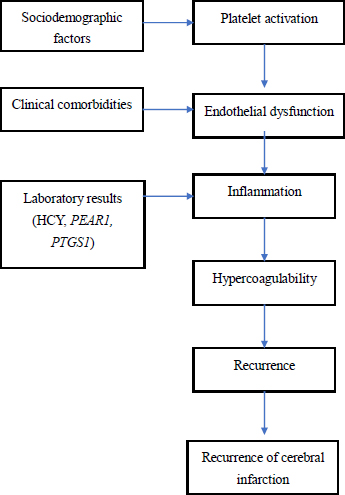
Predictive pathway of cerebral infarction recurrence.
CONCLUSION
The polymorphisms of PEAR1 and PTGS1 genes and the level of HCY have been found to be independent risk factors for the recurrence of cerebral infarction, which can improve the risk sharing of patients with cerebral infarction. Combined with the prediction model (AUC=0.721), a moderately accurate recurrence risk assessment tool has been provided. However, there is a need for prospective, multicenter, and functional studies to validate the findings of this study before clinical application, including patient risk assessment, formulation of individualized secondary prevention strategies, strengthening of the follow-up, and intervention for high-risk patients, in addition to cost-effectiveness analyses.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contribution to the paper as follows: M.H.M.S.: Study conception and design; N.Y.: Data collection; G.Y.: Writing of the paper; R.M.G.: Writing, review, and editing. All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| 95% CI | = 95% Confidence interval |
| AR | = Aspirin resistance |
| AUC | = Area under the curve |
| CI | = Cerebral infarction |
| COX | = Cyclooxygenase |
| HCY | = Homocysteine |
| OR | = Odds ratio |
| PEAR1 | = Platelet endothelial aggregation receptor 1 |
| PTGS1 | = Prostaglandin endoperoxide synthase 1 |
| ROC | = Receiver operating characteristic |
| RR | = Relative risk |
| SNP | = Single-nucleotide polymorphism |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study has been approved by the ethics committee of Beihua University, Malaysia Affiliated Hospital (reference no.: 2024005).
HUMAN AND ANIMAL RIGHTS
All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
AVAILABILITY OF DATA AND MATERIALS
All the data and supporting information are provided within the article.
FUNDING
This study was conducted as part of the Jilin Provincial, Malaysia, Department of Education Project (project no. JJKH20250825KJ).
ACKNOWLEDGEMENTS
The authors extend their sincere gratitude to the staff at the Affiliated Hospital of Beihua University, China, for their invaluable assistance in data collection and technical support.


