All published articles of this journal are available on ScienceDirect.
Virtual Screening for Finding Novel COX-2 Inhibitors as Antitumor Agents
Abstract
The cyclooxygenase-2 (COX-2) enzyme binds to arachidonic acid resulting in the release of metabolites that induce pain and inflammatory responses. Recent studies have shown that strong COX-2 expression is highly correlated with increased tumor risk. Therefore, the development of potent COX-2 inhibitors to relieve pain and treat cancers requires further investigation. We used virtual screening to find three COX-2 inhibitors (Phar-95239, T0511-4424 and Zu- 4280011) from a huge zinc database containing 2000000 compounds. The effects of the compounds on COX-2 were compared to those on COX-1 using a colorimetric COX (ovine) screening assay kit. The selectivity index, the ratio of IC50 for COX-1 inhibition to that of COX-2, calculated were MTT assay was used to evaluate the cytotoxic activity of the compounds using different dilutions. The IC50 values were calculated. Based on the results of the MTT assay, the IC50 values for compounds Phar-95239, T0511-4424 and Zu-4280011 were 178.52, 143 and 97.61 µM, respectively, and the selectivity indices of the compounds were 11.36, 12.20 and 20.03, respectively. These results indicated a relationship between the selectivity index and anticancer activity. Zu-4280011 displayed the highest selectivity index and the best results in the MTT assay among selected componds.
INTRODUCTION
The use of nonsteroidal anti-inflammatory drugs (NSAIDs) for the treatment of inflammation and pain is often accompanied by adverse gastrointestinal and renal side effects [1]. The anti-inflammatory activities of these drugs are mediated by the inhibition of cyclooxygenases (COXs), which catalyze the bioconversion of arachidonic acid to prostaglandins[2]. However, the inhibition of COXs may lead to undesirable side effects. Currently, it is well established that there are at least two COX isozymes, COX-1 and COX-2 [3]. The constitutively expressed COX-1 isozyme is produced in a variety of tissues and appears to be important for the maintenance of physiological functions such as gastroprotection and vascular homeostasis. On the other hand, the COX-2 isozyme is induced by mitogenic and proinflammatory stimuli, suggesting the involvement of this isozyme in inflammatory processes. Therefore, the selective inhibition of COX-2, but not COX-1 is useful for treating inflammation and inflammation-associated disorders. COX-2 inhibitors also have lower gastrointestinal toxicities than other NSAIDs [4]. Recent studies have shown that the progression of Alzheimer’s disease is reduced among some users of NSAIDs. Chronic treatment with selective COX-2 inhibitors may slow the progress of Alzheimer’s disease without causing gastrointestinal damage [5, 6]. Therefore, selective COX-2 inhibitors have been developed as a new generation of NSAIDs with diminished GI side effects. However, rofecoxib and valdecoxib, which are highly selective COX-2 inhibitors, have been withdrawn from the market due to an increased risk of cardiovascular complications. COX-2 mediates the biosynthesis of prostacyclin which is a vasodilator and an inhibitor of platelet aggregation. The indirect inhibition of prostacyclin production by selective COX-2 inhibitors might account for their adverse cardiovascular effects [7, 8].
In addition to the role of COX-2 in rheumatoid arthritis and osteoarthritis, COX-2 expression is triggered by inflammation and carcinogenesis. [4] COX-2 is overexpressed in many solid tumors such as colon [9], breast [10], prostate [11], liver [12] and lung [3] cancers. COX-2 inhibition in vitro using specific COX-2 inhibitors has demonstrated that COX-2 is a potential target for novel cancer therapies [13, 14]. Research attempts to discover selective COX-2 inhibitors have produced many classes of compounds such as coxibs that possess the desired selectivity. For this reason, novel scaffolds with high selectivity for COX-2 inhibition must be identified and evaluated for their anticancer effects [15].
Virtual screening (VS) is a computational technique that is used in drug discovery research and involves the rapid in silico assessment of large libraries of chemical structures to identify structures that will most likely bind to a drug target such as a protein receptor or enzyme [16, 17].
As part of our program to discover novel types of selective COX-2 inhibitors, we now report a methodology to find a new group of COX-2 inhibitory compounds and evaluate their anti-inflammatory and anticancer activities [18].
MATERIALS AND METHODOLOGY
In Silico Studies
Initially, we downloaded approximately 2000000 compounds from the ZINC database (http://zinc.docking.org/). Approximately 12000 compounds with a structure similar to that of the selective COX-2 inhibitor, celecoxib, were selected using ChemOffice (www.cambridgesoft.com). A structure-based virtual screening approach was used to search for new COX-2 inhibitors. The virtual screening was performed using the crystal structure of mouse COX-2 bound to SC-558, which is a COX-2-selective inhibitor (PDB code 6COX) [19]. The ZINC database (using 12000 commercially available compounds) was computationally screened based on binding to the active site of COX-2 using the AutoDock 3.5 program [20]. The calculation by virtual screening took approximately 2.5 days to run on Linux cluster including six Pentium 3-GHz CPUs.
The 40 top-scoring compounds were visually inspected for the plausibility of their predicted binding modes. We selected three of the best compounds with lower docking energies for biological testing.
In Vitro Cyclooxygenase (COX) Inhibition Assay
The ability of the test compounds to inhibit ovine COX-1 and COX-2 was determined using a colorimetric COX (ovine) inhibitor-screening assay (Cayman Chemical, Item Number 760111) that utilizes the peroxidase component of cyclooxygenase. The peroxidase activity is colorimetrically assayed by monitoring the appearance of oxidized N,N,N',N'-tetramethyl-p-phenylenediamine (TMPD) at 590 nm [1].
Cell Line and Culture
The human breast cancer cell line, MCF-7, was obtained from the Pasteur Institute (Tehran, Iran). Cells were maintained at 37°C in a humidified atmosphere (95%) containing 5% CO2. Different cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% v/v fetal bovine serum, 100 units/ml of penicillin and 100 μg/ml of streptomycin.
MTT Assay
The MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenylte-trazolium bromide] method was performed to assess the cytotoxicity of different compounds [21,22]. Briefly, 10000 MCF-7 cells were seeded into 96-well plates and cultured overnight. The cells were treated with different concentrations of the test compounds (1-200 µM). After 24h, 20 μl of MTT was added to each well to give a final concentration of 0.5 mg/ml, and the cells were incubated for 3 h at 37°C. The formazan dye was solubilized with DMSO. The absorbance was measured at 545 nm (630 nm as a reference) in an ELISA reader (Start Fax-2100, UK).
RESULTS
In Silico Studies
We used virtual screening to select three compounds (Phar-095239, Zu-4280011, T0511-4424) with lower docking energies. Celecoxib and SC-558 were used as positive controls (Fig. 1).
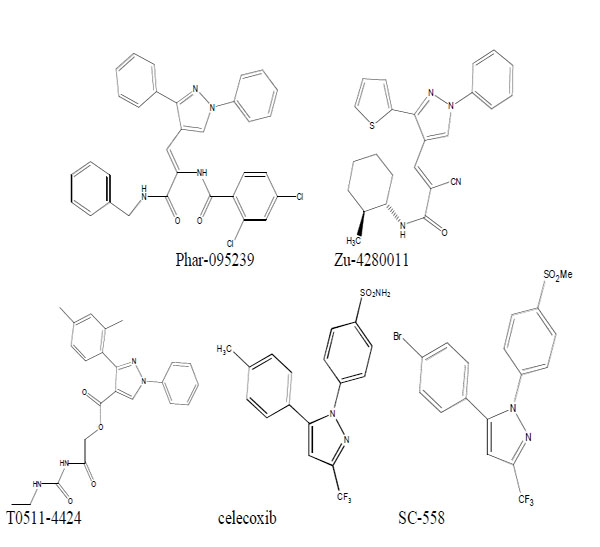
Chemical structure of the test compounds (Phar-95239, T0511-4424, Zu-4280011) and positive controls (celecoxib and SC-558).
Docking view of compounds Phar-095239 (a), T0511-4424 (b), Zu-4280011 (c), and celecoxib (d) at the COX-2 active site has been shown in Fig. (2). The docking energy of the compounds (approximately -14 kcal/mol) was higher than that of the positive controls: SC-558 (-10.38 kcal/mol) and celecoxib (-8.02 kcal/mol).
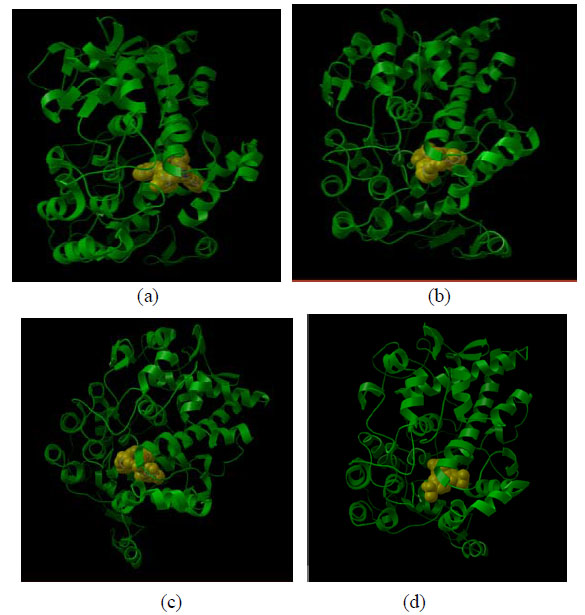
Docking view of compounds Phar-095239 (a), T0511-4424 (b), Zu-4280011 (c), and celecoxib (d) at COX-2 active site.
Cyclooxygenase Inhibition Studies
The test compounds (Phar-95239, T0511-4424, Zu-4280011) were subjected to in vitro cyclooxygenase (COX) inhibition assays (Table 1). Concentration-COX-2 activity inhibition curve of three test compounds and celecoxib are shown in Fig. (3). The IC50, the concentration of compounds that provided 50% inhibition of COX-2 and COX-1, was calculated. IC50s of the test compounds (Phar-95239, T0511-4424, Zu-4280011) and celecoxib for inhibition of COX-1 were 9.32, 8.42, 15.23 and 13.02µM , respectively. IC50 values for COX-2 inhibition were found to be 0.82, 0.69, 0.76 and 0.49µM, respectively. Selectivity indices or the ratio of IC50 for COX-1 inhibition to that of COX-2 of the test compounds and celecoxib were 11.36, 12.20 , 20.03 and 26.57, respectively. The results are shown in Table 1.
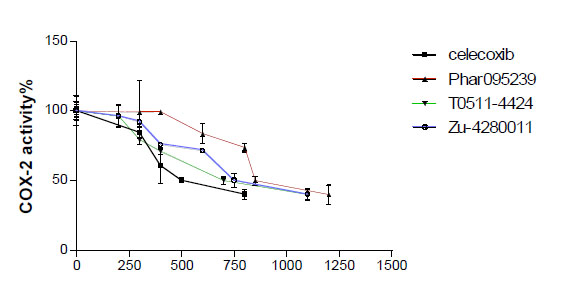
Concentration-COX-2 activity inhibition curve of three test compounds and celecoxib.
Inhibitory Effects of the Test Compounds on COX Enzymes and MCF-7 Cells Proliferation
* This code is used to access compounds in zinc database.
**
The compound Zu-4280011 displayed a high selectivity index of 20.03 near that of celecoxib (26.57) and inhibited COX-2 activity to an extent similar to that of celecoxib.
Effects of the Test Compounds on Cell Viability
Different concentrations of the test compounds (1-200 µM) were used in the MTT assay on MCF-7 cells. The compounds decreased cell viability in a concentration dependent manner (Fig. 4). IC50 for MCF-7 growth inhibition of celecoxib and test compounds (Phar-95239, T0511-4424, Zu-4280011) were 87.69, 178.52, 143 and 97.61, respectively (Table 1). Zu-4280011 was the most potent compound second to celecoxib.
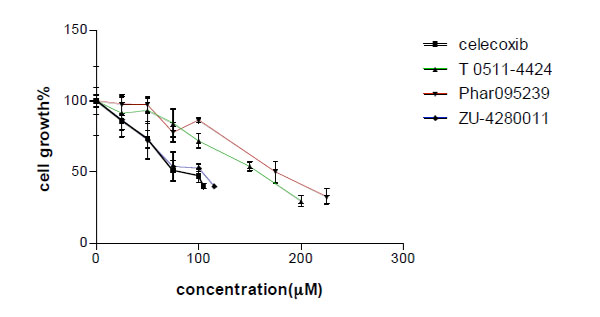
Concentration-cell growth inhibition curve of three test compounds and celecoxib. Cell growth inhibition was measured by the MTT assay.
DISCUSSION AND CONCLUSION
We used virtual screening to find three COX-2 inhibitors from a huge zinc database (2000000 compounds). According to docking studies, we predicted that IC50 values of these compounds for inhibition of COX-2 were less than celecoxib. Although the calculated IC50 values were not exactly what we expected, finding three potent COX-2 inhibitors among a large database was a remarkable achievement, thanks to virtual screening.
We compared the COX-2 IC50 values with MTT assay results (Table 1). We concluded that the order of the inhibitory effects of compounds on MCF-7 cell line proliferation did not correlate with their inhibitory effect on COX-2 enzyme. The order of IC50 values for COX-2 inhibition was: celecoxib < T0511-4424 < Zu-4280011 < Phar-095239 and the IC50 values for MTT assay was: celecoxib < Zu-4280011 < T0511-4424 < Phar-095239.
Our results demonstrated a relationship between the order of selectivity index and MTT assay results. As the selectivity index values increased, the IC50 values in the MTT assay decreased. In other words, more selective compounds had more growth inhibitory effects. The order of selectivity index was celecoxib >Zu-4280011 > T05114424 > Phar-095239 and that of IC50 values for MTT assay was Phar-095239 >T0511-4424 >Zu-4280011> celecoxib. Among these compounds, Zu-4280011 displayed the highest selectivity index (20.03) and the best results in the MTT assay (97.61µM). It can be a candidate for further studies as one potential effective anticancer agent.
We concluded that the compounds’ abilities to inhibit proliferation in cancer cells did not significantly correlate with their inhibitory effects on the COX-2 enzyme. The results of the MTT assay demonstrated a relationship between the selectivity index and anticancer activity. As the selectivity index increased, the growth inhibitory effects of the tested compounds decreased.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENT
The authors gratefully acknowledge the funding provided by the Vice Chancellor of Research, Mashhad University of Medical Sciences for this project The results described in this paper are part of a PharmD thesis.


